Adaptation of jumping gene boosts CRISPR-Cas gene-writing technology to new heights
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

As of December 30th, 2021, the Online Mendelian Inheritance in Man compendium indicates that mutations in 4,561 genes are linked to human genetic disorders. Looking at the underlying mutation types, about 22% of reported human mutations involve deletions.
State-of-the-art gene-editing technologies that are being developed for clinical applications have only limited potential to effectively and efficiently correct deletions in genomic DNA.
Two seminal scientific reports were recently published describing new gene-editing technologies that aim to fill this gap (1, 2). In one of these reports, published in Nature Communications, a research team led by Professor Marc Güell at the Translational Synthetic Biology lab at Pompeu Fabra University revealed a new gene-writing tool called ‘FiCAT’- for find, cut and transfer, which was specifically developed to repair sizable defects in genomic DNA.
Building on experience in human genetics, molecular and synthetic biology
»The development of FiCAT gene-editing was one of the first projects we initiated when starting the lab in 2017,« says Professor Marc Güell. »Much earlier in my career, I had realised that my scientific interest was focused on technology development. My mentor would always joke that I was paying more attention to the methods than to the actual outcome of my experiments. It was a wonderful opportunity to continue my career as a post-doctoral researcher in the renowned tech lab of George Church at Harvard Medical School. I felt like a kid in a candy store! There, I started working on developing CRISPR methods for genome recoding to make xenotransplantation a clinical reality - research that is now continued by a company called eGenesis. From Boston, I came back to Barcelona to establish my lab with a focus on precise gene-writing in mammalian systems and engineering of the skin microbiome.«
Dr. Avencia Sánchez-Mejías takes over: »After my PhD in clinical genetics at the University of Seville, I performed postdoctoral research in molecular oncology in Miami and Singapore. Basic research, but always aimed at finding new treatment options for patients with cancer. When I returned to Spain, Marc had just launched the lab and presented the research interests on precise gene-editing. I was super inspired. During my PhD, I had counselled couples carrying various genetic conditions. We also did preimplantation diagnosis of early embryos for affected couples, but this was still very far from a reality where gene therapy could make a difference for children with a genetic disease. By developing gene-editing technology for clinical use, we could really start making a difference for these patients with unmet needs. In February 2022, it will be over four years since we began our work on FiCAT.«
Defining gene-writing research challenges
Established gene-editing techniques that rely on DNA templates for introducing new sequence information by homology-directed repair (HDR) are hampered by low efficiencies and become even less effective when insert sizes increase. Base editors are not suited to correct even small deletions, and while standard prime editing can correct small deletions, new technology is needed to engineer the precise delivery of large DNA coding regions.
»The basic idea behind FiCAT is to combine the genomic precision and DNA targeting ability of Cas9 with highly effective DNA transfer activity. This combined power would create a gene-editing tool capable of correcting genomic deletions or precisely rewriting large stretches of genomic DNA at once,« says Professor Güell.
Previously, homology-independent targeted integration (HITI), relying on the non-homologous end joining of the double-strand break repair process, was successfully used to specifically integrate DNA fragments of several kilobases in length, and could potentially work to insert missing exons. However, this technology may not be capable of delivering large DNA fragments required for restoring gene function in diseases as Duchenne muscular dystrophy (dystrophin DMD cDNA ~14 kb) or cone-rod dystrophy (ABCA4 cDNA ~6.8 kb), for example. To circumvent this delivery challenge, the team turned to the piggyBac transposase, as Dr. Sánchez-Mejías explains:
»In bacteria, it had been shown that precise gene delivery could be performed by combining CRISPR-Cas with transposon elements: the natural ‘jumping genes’ that exist in many species and can excise themselves to relocate to other regions in a genome. However, CRISPR programmable transposon technology did not work in mammalian cells. This was our challenge, adapting CRISPR-Cas and the piggyBac transposase to work together.«
Teamwork makes gene writing better
To make this project work, a synergistic research team was assembled from different fields of expertise including classic genetics, evolution, computational and synthetic biology, and protein engineering. »The first step was to make a protein-protein fusion between Cas9 and the piggyBac transposase and create a fluorescent reporter system that lights up the cells after successful CRISPR-guided payload delivery.» (Figure 1A).
»piggyBac fused to the carboxy terminus of nuclease active Cas9 showed the best transposon DNA delivery capacity. However, specificity was still a concern, as the endogenous DNA binding capacity of the transposase would also result in transposon insertions at other genomic loci,« explains Dr. Sánchez-Mejías.
Professor Güell explains how the team addressed this challenge:»To solve this specificity issue, we needed to separate the functionality of the fused Cas and piggyBac proteins. Since the programmable Cas9 is used to find the genomic target and create a double strand break for payload insertion, the native DNA-binding affinity of transposase is no longer required - and even undesirable, as this results in off-targets. So, we engineered piggyBac to lose its DNA-binding affinity. We did this based on previously identified mutations and protein modelling using a structural prediction algorithm to identify critical residues by comparing DNA binding domains of related proteins.«
Dr. Sánchez-Mejías explains how removing piggyBac’s DNA-binding affinity gives an additional advantage. »A result of these modifications is that the TTAA transposition recognition and insertion motif which is normally required for DNA integration is no longer needed at the genomic target locus. The transposon DNA payload with required inverted repeats at both ends is nicely positioned in the targeted double strand break created by Cas9. At this location, our experiments do reveal frequent small indels as a likely result of DNA end processing mechanisms in combination with transposition.« (Figure 1B). »However, importantly, the inserted DNA payload separates the guide RNA (gRNA) recognition sequence from the protospacer activation motif, preventing cyclic Cas9 activity. And similarly, the absence of a TTAA motif in the genomic target also means that payload delivery is irreversible, as the transposase is incapable of excising the inserted DNA,« adds Dr. Sánchez-Mejías.
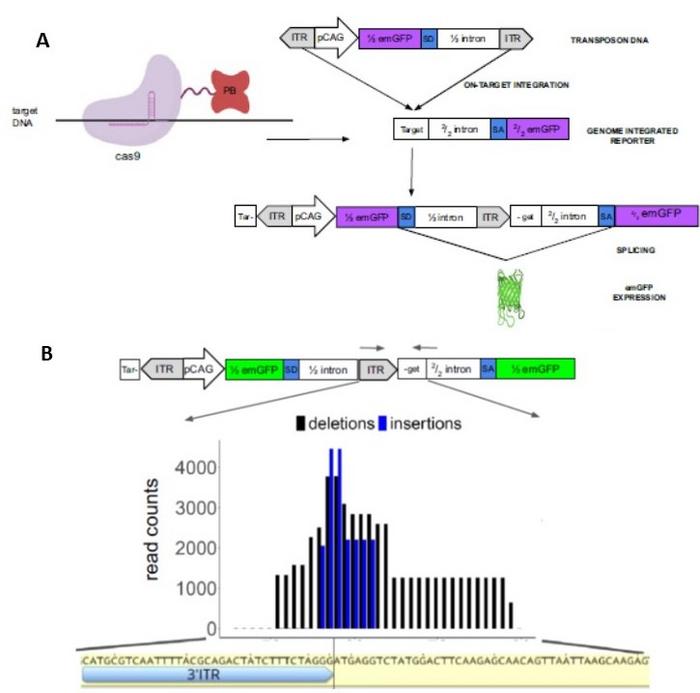
FiCAT performance and directed evolution
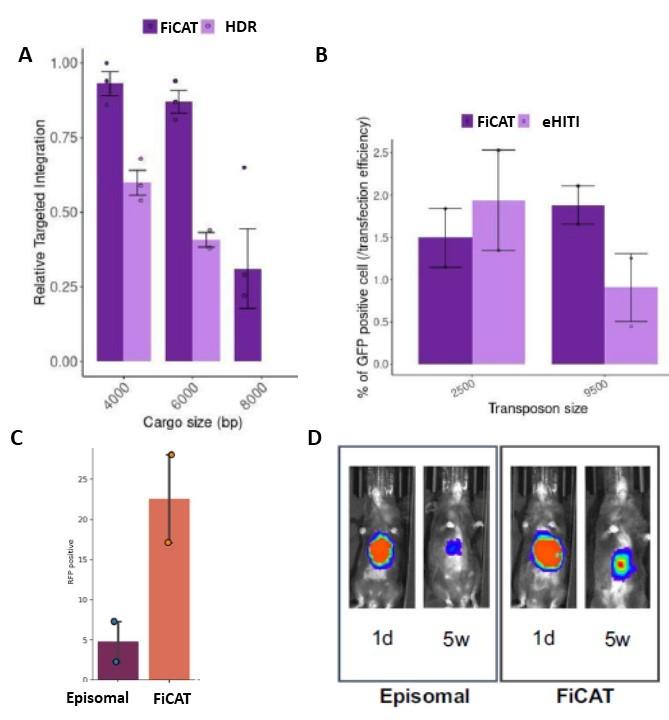
Having established a proof of principle of the find, cut, and transfer (FiCAT) gene-editing technology, experiments were performed to determine its effectiveness. In their Nature Communications paper published December 3rd, 2021, the team reported that FiCAT resulted in two-fold higher delivery efficiencies in comparison to methods involving HDR or enhanced HITI to create targeted DNA insertions, achieving insertion up to 8 kb with high precision (Figure 2 A,B). After some tweaks and optimisation, FiCAT on-target insertion efficiency could be pushed up to 22% of cultured cells (Figure 2C), with early experiments suggesting off-target risks are manageable and comparable to spCas9.
A potential additional advantage of FiCAT is that circular DNA templates can be used, in contrast to the linear DNA fragments required by HDR or HITI, which are more likely associated with increased genome toxicity and uncontrolled insertion risks.
»Building on the tissue culture experiments, we investigated whether FiCAT-guided transposition could also be used in vivo,« says Professor Güell. »Transposon plasmids or minicircles combined with gRNAs and mRNA encoding the Cas9-piggyBac enzyme were injected into the bloodstream of mice either by hydrodynamic injection or enclosed in in vivo JetPei® particles. We targeted the transposon delivery into the safe harbour Rosa locus in mouse liver cells, with successful delivery confirmed by a fluorescent reporter protein or luciferase expression. Stable delivery was confirmed by continued transgene expression over a time frame of 4 to 5 weeks«. (Figure 2D).
»We had now established that we had developed a potentially clinical functioning gene-writing tool and we put a lot of effort into optimising FiCAT by directed evolution. Focusing on 17 specific amino acids, we created nearly 200,000 FiCAT variants that were screened in lentiviral combinatorial libraries for efficacy. With that, we were able to reduce the intrinsic DNA binding capacity of piggyBac even more, while increasing the proficiency of the DNA payload for insertion.«, says Professor Güell.
Joey Riepsaame PhD, Head of Genome Engineering Oxford at Sir William Dunn School of Pathology, University of Oxford, who was not involved in the current study sees FiCAT as a potential major advance within gene editing:
»The find, cut and transfer (FiCAT) system described by Maria Pallarès-Masmitjà and colleagues in Nature Communications nicely complements the work by Hew et al. who in 2019 showed for the first time that catalytically inactive Cas9 (dCas9) fused to a piggyBac transposase (PBase) mutant can efficiently install large plasmid-derived DNA cargo located between two inverted terminal repeat (ITR) sequences into mammalian genomes. Although efficient, this dCas9-PBase hybrid had a tendency to integrate its transposon cargo into random TTAA piggyBac insertion sequences scattered throughout the genome, resulting in undesired edits. By replacing dCas9 with catalytically active Cas9 in this system, Pallarès-Masmitjà et al. seem to have solved this issue and show for the first time that FiCAT specifically installs transposon-derived DNA cargo up to 9.5 kb at the gRNA-defined target site. Unlike its predecessor, FiCAT-mediated transposition seems to be gRNA-dependent because the DNA cargo is specifically inserted at/near the Cas9 cleavage site, instead of nearby or random TTAA insertion sequences. Therefore, FiCAT could be a major step forward for the specific and stable introduction of large DNA sequences into user-defined target sites within mammalian genomes.«
Path to clinical FiCAT applications
Having established the potential clinical value of FiCAT, Dr. Sánchez-Mejías explains what is next for future development:
»From the beginning, it was important that this technology would be made available for the patients in need. We thought that the best way to push the clinical development process forward was to create a spin-off company, Integra Therapeutics. It was clear to us that involvement of the pharmaceutical industry was essential to take FiCAT from the lab into the clinic. We were fortunate to find a very helpful business development environment in Barcelona that provided financial support for intellectual property protection, and knowledge about technology transfer, strategic planning and how to attract essential venture capital that we needed to further develop FiCAT for therapy.«
At this time, research developments at Integra Therapeutics are not yet focused on specific patients with specific genetic diseases. »We are still really centered on the technological platform itself. It is crucial that we precisely understand what FiCAT can bring to the gene-writing field,« says Professor Güell, who continues: »In addition, we need to focus on targeting cells and tissues and which delivery systems to implement. This all has to be validated in pre-clinical models before we can actually start clinical trials with patients.«
In this context it should be noted that FiCAT gene-writing applications are not just restricted to correcting genetic deletions. FiCAT is also suited to the development of new immune cell therapy against cancer, as Professor Güell elaborates: »It may facilitate the effective delivery of chimeric antigen receptor (CAR) cassettes into safe harbour loci. Also, FiCAT holds clinical promise by enabling targeted insertion of cDNA-based mini-genes.« This strategy could potentially compensate different loss of function mutations in many patients with the same dysfunctional gene, avoiding the need to design tailored gene-editing strategies for each unique patient.
Dr. Sánchez-Mejías takes the last word: »We really see FiCAT as a very versatile asset that has great potential to actually treat different genetic conditions. So, although we are not aiming for specific patients yet, we are working towards clinical applications of FiCAT, and that is really exciting.«
Henri van de Vrugt is a molecular biologist and Chief Scientific Officer at New Haven Biosciences Consulting, US, and Senior Scientist & Advisor at the Dept. of Human Genetics, Amsterdam UMC, the Netherlands.
Cited literature:
1. Pallarès-Masmitjà, M., Ivančić, D., Mir-Pedrol, J. et al. (2021) Find and cut-and-transfer (FiCAT) mammalian genome engineering. Nat Commun 12, 7071. https://doi.org/10.1038/s41467-021-27183-x
2. Anzalone, A.V., Gao, X.D., Podracky, C.J. et al. (2021) Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat Biotechnol. https://doi.org/10.1038/s41587-021-01133-w
3. Hew, B.E., Sato, R., Mauro, D. et al. (2019) RNA-guided piggyBac transposition in human cells. Synthetic Biology. https://doi.org/10.1093/synbio/ysz018
Tags
ArticleInterviewNewsFind and cut-and-transfer (FiCAT)Integra Therapeutics
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







