Considerations when choosing CRISPR genome editing agents

CRISPR-Cas9 gene editing has been around for less than a decade, but it has ignited a blast of human creativity and ingenuity. The last eight years have seen discoveries of many new naturally occurring CRISPR systems, engineering of original Cas variants and invention of new genome editing tools. The result is a vast and varied arsenal of tools, each with their strengths and limitations, and the question is how to pick the best tool for your desired target?
Help has just arrived: a new review in Nature Biotechnology from experts in the Liu Lab. They bring overviews, flowcharts and tables to help compare and choose the right tool for your edit. From asking what type of edit, and what tools can target the desired sequence to more nuanced but important considerations such as cell type, method of delivery and propensity for off-target events.
The researchers point to four classes of CRISPR tools for genome editing: nucleases, base editors, transposases and prime editors, and discuss each one.
We spoke to Andrew Anzalone, who authored the review with Luke Koblan and David Liu, about some of the key points and considerations for each of the four classes.
Nucleases
The review focuses on the most important class II system nucleases - the Cas9 type and Cas12 type nucleases. Very briefly, both nucleases are guided by guide RNA to generate a double-strand break of the DNA after which genome editing results from DNA repair mechanisms. This can be through indels that result in gene disruption, homology-directed repair to install targeted mutations or knock in of a large DNA fragment.
A primary consideration when choosing a nuclease is the protospacer adjacent motif (PAM) requirement. The PAM is a short DNA sequence that must be present near the target DNA site, for example, the classic S. pyogenes Cas9 has an NGG PAM. In the natural bacterial phage defence system, it is a safety mechanism that protects from auto-cleavage, but this imposes a limit on what sequences may be targeted.
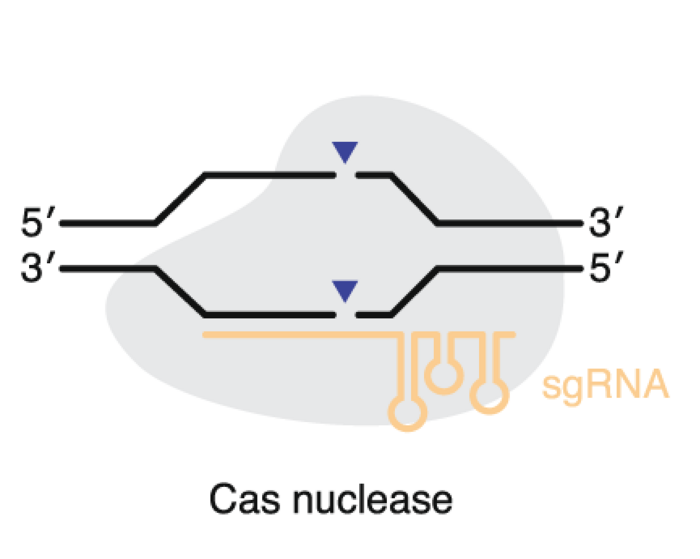
Therefore, to expand the scope of Cas9, a lot of research has focused on engineering the nucleases to have less restrictive PAM requirements, e.g., only NG, or to have a different sequence such as NGA. Engineering, unfortunately, can reduce editing efficiency.
»We've come a long way, and there are amazing tools out there already, but one challenge remains to broaden the scope without reducing the efficiency of the editor,« says Andrew Anzalone.
»We might imagine a future where we have many different Cas9 derivatives, all of which have a different PAM that contains just two nucleotides, and retain the on-target editing efficiency of the wild type nuclease.«
Having found a nuclease with the right PAM, another critical consideration can be high fidelity variants with fewer off-target events. This might not be an issue for a research experiment, but for therapeutic applications, it is particularly important.
Different high fidelity systems have been engineered, but according to Anzalone, most researchers use engineered Cas9 or Cas12 variants with mutations that make them more specific.
»However it is widely recognised that when you reduce the off-target editing efficiency, you're similarly reducing the on-target editing,« says Anzalone.
»It depends on your application - if you're very concerned about off-targets in a clinical setting you might choose to use a high fidelity variant.«
When it comes to clinical applications, another consideration is the size of the editor. For in vivo gene editing, it is a challenge to package everything into the preferred viral delivery vector - AAV (adeno-associated virus) - and discovering or engineeringsmaller Cas variants could open the door to many exciting applications. The S. pyogenes Cas9 is around 1,400 amino acids long, but S. aureus Cas9 is just over a little over 1,000, so Anzalone hopes that even smaller Cas9 variants will be found in the future.
In the review’s supplementary materials, Anzalone and colleagues have compiled handy tables to compare and contrast the different nucleases.
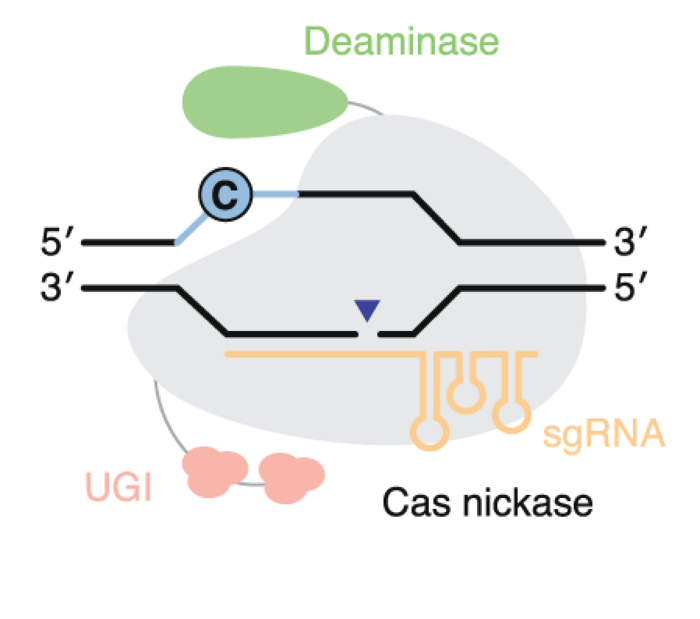
Base editors
Base editors are more complex than nucleases. They contain at least two domains: a Cas domain fused to a deaminase domain, and they precisely install targeted point mutations without requiring double-stranded breaks or donor DNA templates, and without reliance on homology-directed repair.
Two main classes exist: C to T base editors and A to G base editors, that can collectively make 4 of the 12 possible point mutations and cover about 30% of human pathogenic variants.
Because of the rich combinatorial possibilities, many different base editors have been designed and tested in vivo and in tissue culture systems.
To help select one from them all, Anzalone et al. have created a handy flow chart.
»It is sort of a decision tree that I had become accustomed to back in medical school - they always make these flowcharts for what you do given certain situations‚« says Anzalone.
The first decision is what edit you are trying to make; either C to T or A to G.
Then, as with nucleases, you consider whether there is a PAM near the target base.
»A huge consideration for base editors is the PAM because the PAM defines where the window is for base editing,« says Andrew Anzalone.
Depending on the particular target a set of Cas9 or Cas12 variants may exist, where the target base is within a specific distance (window) from the PAM.
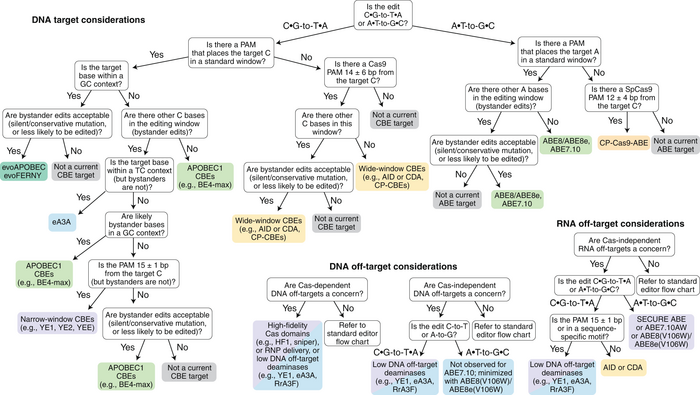
Off-target events are also a consideration for base-editors, and because the deaminase part of base editors may edit matching nucleotides within the window, so-called 'by-stander edits' can be a concern. The edit may result in a silent mutation because of degeneracy in the genetic code, but if not there may be ways to fine tune the editor.
»You can either move the PAM slightly to change the window to favour your edit over others. Or use a different deaminase enzyme that has a preferred sequence context, so that it would favour the edit of interest and disfavour others,« says Andrew Anzalone.
However, the deaminase can also cause a different type of off-target event that happens stochastically. These result from the transient formation of R-loops in DNA - pieces of DNA that expose single-stranded DNA that the deaminase enzymes on the base editors happen to act on.
»There's no doubt that there will continue to be progress made to reduce the off-targets, and it may very well be that editor variants are generated in the future with better off-target editing profiles,« says Anzalone.
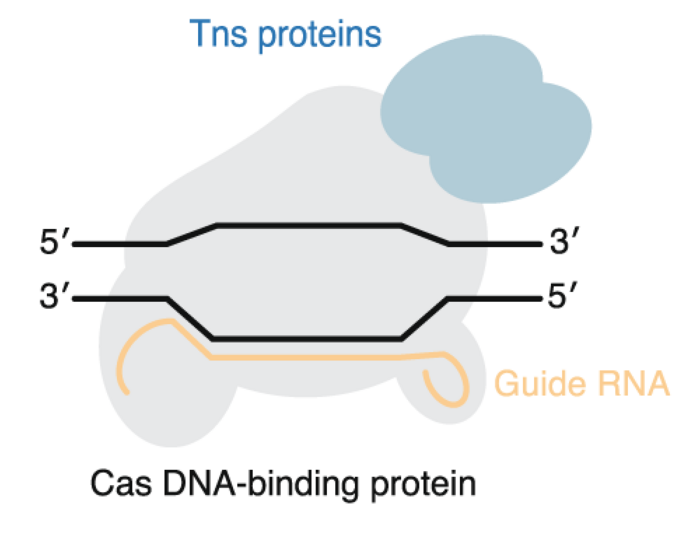
CRISPR transposases
CRISPR transposases are a new tool that were first reported in summer 2019, and despite only being reported for bacterial cells so far, the results have been impressive.
»We all agree that the prospect for the future is very exciting,« says Andrew Anzalone.
The CRISPR transposases are naturally occurring systems that use CRISPR to precisely target a place in the genome where you integrate a cargo piece of DNA such as a fully functional therapeutic gene. Through CRISPR, it may be possible to target the integration to specific safe harbour loci in the genome. Therefore, it would eliminate a safety concern of traditional gene therapy that uses lentivirus which integrates into many different places in the genome. The CRISPR transposases could potentially function like gene therapy, but without the risk of oncogenesis or loss of essential genes.
For now, the system only works in bacterial cells, but looking ahead CRISPR transposases making it to the eukaryotic cell would be a significant milestone.
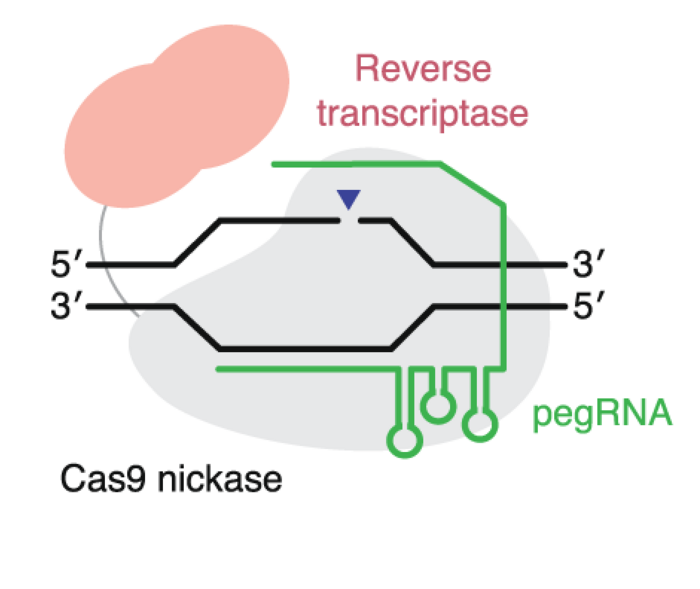
Prime editors
Prime editors are also a very new tool with future potential for human therapy. Prime editors combine a Cas9 nickase and a reverse transcriptase and can make all 12 types of point mutations and make insertions or deletions of up to 50 nucleotides. You can read more about prime editors in this article.
The first encouraging results in mammalian cells have shown prime editing to work in mouse embryos and human induced pluripotent stem cells, but exploration has only just begun.
»I think the most important questions around prime editors are questions about what else you can do with it? Can you make much larger changes? Can you integrate much larger pieces of DNA into the genome? And questions about how do we get prime editing to be more efficient, and more efficient in the therapeutically relevant cell types,« says Andrew Anzalone.
A large part of the prime editing process relies on DNA repair, and research in this area could be fruitful for increasing efficiency.
»I think finding out what proteins and cell factors are involved in prime editing would be really relevant and very useful. If you could predict how to perturb the cell’s DNA repair machinery to favour prime editing, that would be great,« says Anzalone.
As with the exploration of base editors, that has been extremely productive in the past three years, he thinks many areas are open to improvements such as the design of the guide RNA (called prime editing guide RNA or peg RNA) and exploring new variants of the reverse transcriptase.
»I hope that in two or three years there will be a comparable amount of development made with prime editing as has been made for base editing,« says Andrew Anzalone.
Tags
AnalysisArticleOff-targetReagentsBase editorsCRISPR-CasPrime editors
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







