Explainer: What Is Prime Editing and What Is It Used For?
What is prime editing and what is it used for?
Prime editing was described initially by the laboratory of Dr. David Liu in 2019 (1). This powerful tool combines the DNA-scanning and sequence-identification capabilities of the CRISPR-Cas9 system with a reverse transcriptase enzyme, which uses an RNA template to synthesise a new single-strand DNA (ssDNA) sequence and insert it into the DNA.
Prime editors (PEs) can introduce any of the twelve possible types of nucleotide substitutions using the same PE protein configuration (1). This allows greater flexibility than base editing, which requires a different protein configuration for each of the four types of nucleotide substitutions that are feasible to date (see our explainer about base editing for more details).
In addition, PEs can also introduce small-to-medium-sized targeted insertions and deletions with no or very few unwanted modifications at the target site, and virtually no off-target effects (2). This is because PEs, unlike other CRISPR-Cas9-mediated targeted integration approaches, do not generate DSBs as part of their gene-editing process. This greatly reduces the potential for DSB-related chromosomal aberrations and cell toxicity, which are two important limiting factors for safe gene editing.
What are the components of a prime editor?
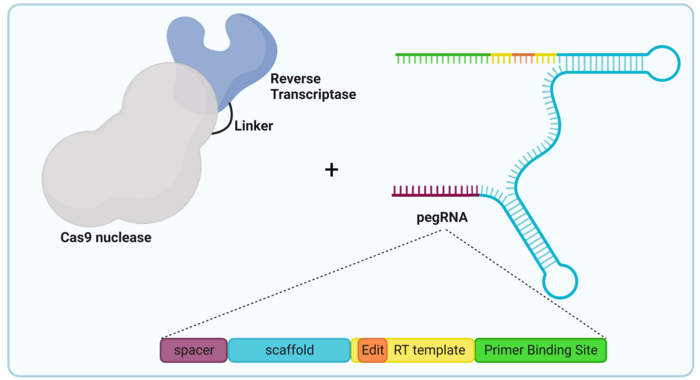
Similarly to base editors, PEs are generated by coupling two separate proteins capable of very specific functions, as illustrated in Figure 1. The components of a typical prime editor include:
- Cas9 nickase (nCas9): This is a version of Cas9 that has been modified through mutations in one of the two main amino acid residues responsible for the DNA cleavage activity of Cas9. Thus, nCas9 is still able to pair with a gRNA and find the DNA sequence complementary to the gRNA spacer but is only able to nick one strand of the DNA.
- Reverse transcriptase (RT): This is a virus-derived enzyme which is capable of reading an RNA sequence and transcribing it into a single-stranded DNA sequence. Most PEs use an RT enzyme derived from the Moloney murine leukaemia virus (MMLV-RT). This RT is fused to nCas9 through a peptide called a linker.
- Prime-editing guide RNA (pegRNA): This is the most important and complex element of any PE, and it defines both the target sequence and the genetic modification to be introduced. A pegRNA consists of four main parts:
- Spacer: This sequence, found at the 5’ end of the pegRNA, targets the whole PE complex to a specific DNA sequence, in the same way that the spacer sequence of a regular gRNA would target Cas9.
- Scaffold: This scaffold is an RNA sequence that folds on itself with a two-dimensional structure that allows it to bind to the nCas9 protein. At the same time, it connects the spacer with the prime-editing template.
- Primer-binding site (PBS): This sequence is found at the 3’ end of the pegRNA and is homologous to the DNA sequence just upstream of the target site determined by the spacer. This homology leads the PBS to anneal to the target DNA and acts as a primer for the RT.
- RT template sequence: This sequence is found immediately after the PBS and serves as a template for the RT to synthesise the DNA sequence that includes the edit and which will be introduced at the target site.
How does prime editing work?
Prime editing is a complex process that involves a nick of the non-complementary strand and the hybridisation of the pegRNA and the nicked strand, which serves as a primer for reverse transcription (Ref. 2, see Figure 2).
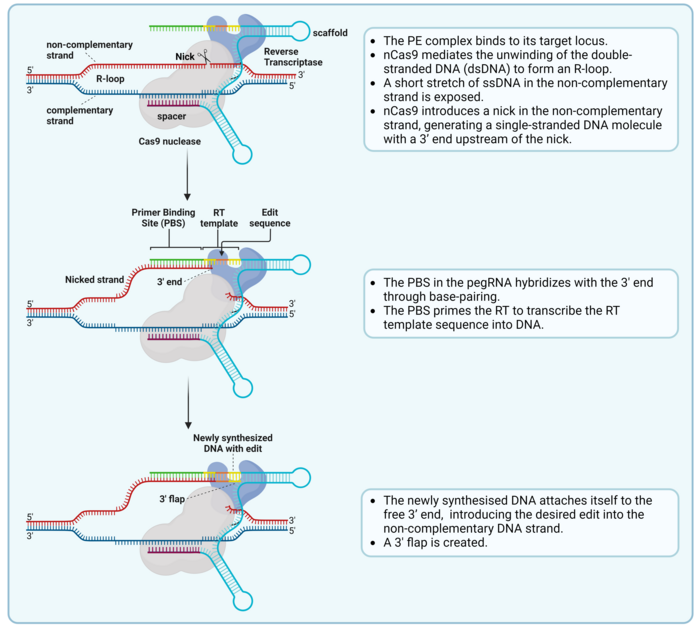
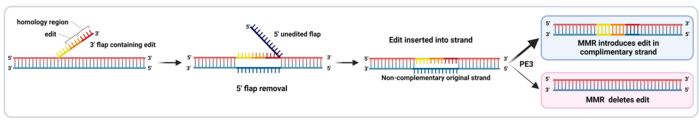
The final part of the RT template sequence includes a region of homology with the DNA sequence downstream of the nick, which allows the 3’ flap to align with the complementary strand (Ref 2, see Figure 3). To make room for this alignment, the 5’ end that was present downstream of the nick in the target sequence is detached from the complementary strand as a 5’ flap and removed by endogenous cellular DNA repair mechanisms. Then, the remaining nick is repaired, and a new double-stranded DNA (dsDNA) molecule is formed (Figure 3).
However, since the introduced edit is only present in the non-complementary strand, the new dsDNA contains two strands with different information in them. This causes incomplete alignment between both strands, also known as a mismatch. This is repaired by the mismatch repair (MMR) machinery, which randomly takes one of the two strands as a template and repairs the other strand to match it. This means that in half the cases, the MMR will reject the edit and re-construct the original sequence. However, a newer PE version called PE3 includes an extra sgRNA, which is used by the PE complex to nick the strand which does not include the edit. This leads to the MMR machinery preferentially using the edited strand as a repair template, which introduces the edit in the other strand and generates the final dsDNA edited sequence (Figure 3). PE3 introduces edits with a 4.2-fold higher efficiency than PE2 (1,2).
How has prime editing been used so far?
Several fascinating therapeutic approaches based on prime editing have been explored all over the world in the last few years. For example, in August 2021, the group of Hyongbum Henry Kim (Yonsei University) published the first report of successful in vivo prime editing both in the retina and in the liver (3), achieving correction of mutations and phenotypes in mouse models of Leber’s congenital amaurosis and hereditary tyrosinemia, respectively. Similarly, the group of Gerald Schwank (University of Zurich) reported in March 2022 that they performed liver-directed prime editing in a mouse model of phenylketonuria, achieving therapeutic reduction of blood phenylalanine (4). The group of David Liu (Harvard University) has also recently reported successful prime editing in the mouse brain, liver and heart (5).
Beyond therapeutic applications, scientists are also exploring the possibility of using prime editing for other applications, including the generation of disease models, screening assays, and very importantly, the generation of crops with desired properties that can facilitate agriculture in the future.
What are the limitations of prime editing?
Despite its advantages over other gene-editing technologies, prime editing presents some limitations that are currently being addressed by scientists all over the world (2,6).
Some of the limitations are similar to those of conventional Cas9 editing approaches regarding targeting and delivery. Thus, already existing Cas9 variants that were developed to solve those issues can be used to construct prime editors able to overcome the same issues. These new PE variants can expand the range of target DNA sequences that can be edited, but they are generally less efficient than the original PEs, which suggests that further optimisation is required.
When it comes to in vivo delivery, the increased size of the PE complex compared to regular Cas9 complicates its delivery. To address this hurdle, the intein system, which is used to package other Cas9 products into adeno-associated viral vectors, has also been used for in vivo delivery of base editors. Other authors have also used ribonucleoprotein complexes embedded in virus-like particles, which overcomes packaging issues and guarantees short-term expression of the PE (7). However, due to the nature of prime editing, its other limitations are quite different from those of other gene editing approaches (2):
- Efficiency: So far, the most relevant limitation of prime editing has been its relatively low efficiency. This is because prime editing consists of several key steps that need to occur in concert, and the efficiency of each step significantly impacts overall editing efficiency. For this reason, improving the efficiency of every step has been the main target of scientists working in the prime-editing field. To this end, new PE variants with improved nuclear localisation, codon optimisation, additional DNA-binding domains and even altered architectures have been developed and shown to improve prime-editing efficiency.
- pegRNA design: In contrast to base editing, where the design of the base-editor complex determines the edits to be introduced, in prime editing the edits are determined by the pegRNA. Thus, the pegRNA is the most essential part of the PE, and its design can have an immense impact on prime-editing efficiency. Thus, testing and comparing pegRNAs with different PBS and RT template sequence lengths has been shown to be crucial to achieving efficient editing. Additionally, introducing structural modifications to the 3’ end of the pegRNA has been shown to protect it from degradation by ribonucleases, which significantly improves editing efficiency. Similarly, other modifications aimed at improving pegRNA transcription and stability also have an effect.
- INDEL generation by PE3: An important limitation of PE3 is that the nick generated in the non-edited strand can, in rare cases, coincide in time with the nick introduced by the pegRNA in the edited strand. This can result in a DSB and thus the generation of INDELs. While the occurence of INDEL generation is significantly lower using PE3 than using CRISPR-Cas9-mediated targeted integration approaches, it is still a significant concern. To address this, a new PE derivative called PE3b, where the sgRNA used to cut the non-edited strand can only recognise this strand after the integration of the 3’ flap, has been developed (1). This PE3b strategy completely negates INDEL generation.
- MMR antagonism: Researchers have discovered that the high efficiency of the MMR system can antagonise prime editing, significantly reducing its efficiency and stimulating INDEL by-products. Strategies targeted to avoid the activity of MMR at the edit site can reduce the formation of INDELs and increase prime-editing efficiency. These strategies include the introduction of silent mutations in the RT template sequence, which increases the mismatches between the edited and non-edited strand, as well as the fusion of the PE to a dominant negative variant of an MMR protein, which will occupy the edit space and inhibit other, active MMR proteins from accessing it. These approaches increase prime-editing efficiency up to sevenfold, but they require the co-delivery of a third protein, which further complicates the aforementioned delivery issues.
- Insert size: While early PEs could only produce relatively short insertions (up to 100bp) and deletions (up to hundreds of bp), newer PEs have been developed that can introduce larger edits. To increase the size of prime editing-mediated edits, scientists have explored the use of pairs of pegRNAs on opposite strands, pegRNAs that cause recombination between distal loci (up to 10kB), and the combination of prime editing with recombinase-mediated integration.
Stay tuned for future explainers about various gene-editing technologies. In the meantime, you can find all of our coverage on prime editing right here.
Cited literature:
1. Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019. 576(149–157).
2. Chen, P. J. & Liu, D. R. Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 2023. 24(161–177).
3. Jang, H. et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 2022. 6(181–194).
4. Böck, D. et al. In vivo prime editing of a metabolic liver disease in mice. Sci. Transl. Med. 2022. 14(eabl9238).
5. Davis, J. R. et al. Efficient prime editing in mouse brain, liver and heart with dual AAVs. Nat. Biotechnol. 2023. doi:10.1038/s41587-023-01758-z.
6. Zhao, Z. et al. Prime editing: advances and therapeutic applications. Trends Biotechnol. 2023. 41(1000–1012).
7. Haldrup, J. et al. Engineered lentivirus-derived nanoparticles (LVNPs) for delivery of CRISPR/Cas ribonucleoprotein complexes supporting base editing, prime editing and in vivo gene modification. Nucleic Acids Res. 2023. 51(18):10059–10074.
Manel Llado is a gene editing expert and science writer/communicator based in Vienna, Austria
We acknowledge Dr. Jakob H. Haldrup (Radcliffe Department of Medicine, Oxford) for advice and input.
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University








