Innovative Biosensor Uses CRISPR-Cas12a and PER to Quantify Pathogenic E. coli
Pathogenic bacteria in contaminated food is a serious problem to public health. One of the most common foodborne pathogens is E. coli O157:H7.
Although most frequently found asymptomatically in ruminant animals, E. coli O157:H7 is the leading cause of haemorrhagic diarrhoea, haemorrhagic colitis, and haemolytic uremic syndrome in humans. In the USA, the strain causes approximately 2,100 hospitalisations and up to 20 deaths per year. The infections are often associated with ingesting contaminated hamburgers, milk, water, fruit, and vegetables, so it is vital to ensure early detection of this and other pathogenic bacteria in human foods.
Traditionally, E. coli O157:H7 has been detected with labour-intensive and time-consuming culturing approaches. These are often based on the strain's inability to ferment sorbitol on Sorbitol-MacConkey (SMAC) agar. As a result, O157:H7 colonies appear transparent, while colonies of the usual sorbitol-fermenting E. coli strains appear red. More rapid detection methods utilising PCR, fluorescence and ELISA can cut detection times from days to several hours. Still, their application is limited by a relatively high cost and the need for specialised equipment and trained technicians. To solve these problems, Wensen Liu and Jiayu Wan from Changchun Veterinary Research Institute in China have developed an ultrasensitive electrochemical biosensor for point-of-care testing of pathogenic E. coli O157:H7.
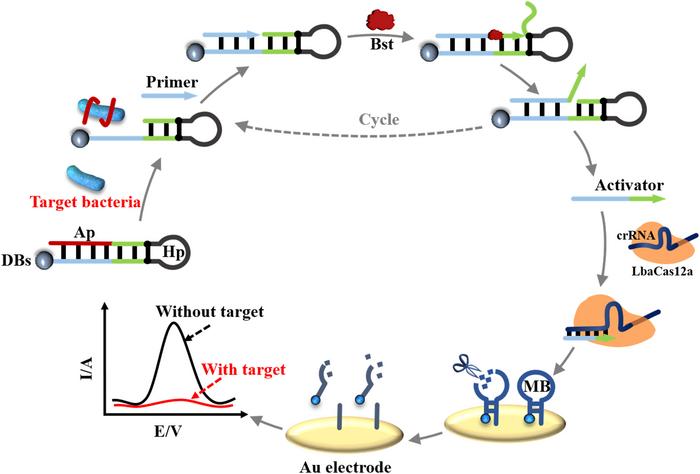
The method was published in August in Sensors and Actuators B: Chemical, and while it utilises unspecific, collateral CRISPR-Cas12a activity to release an electrochemical signal, it does not require nucleic acid extraction. Instead, it exploits functional DNA aptamers that specifically bind to E. coli O157:H7 if the pathogenic strain is present. Binding unlocks a primer exchange reaction (PER) and initiates the production of single-stranded DNA (ssDNA). The ssDNA subsequently unleashes CRISPR-Cas12a activity and leads to an electrochemical signal detected by differential pulse voltammetry (DPV).
Aptamers block detection in the absence of target bacteria
The method allowed for quantification of E. coli O157:H7 in a linear range from 10 to 106 CFU·mL-1 with a detection limit of 19 CFU·mL-1. Previous electrochemical techniques have employed aptamers or antibodies as recognition elements for E. coli O157:H7 or other pathogenic bacteria in combination with a range of detection assays. The new method described by Liu and Wan, however, is the first to combine aptamers, Cas12a and PER, and it has one of the lowest detection limits and also one of the broadest quantification ranges, spanning six orders of magnitude.
The PER mechanism is central to the new detection method. In 2018, Peng Yin from Harvard University introduced it for the programmable autonomous synthesis of ssDNA using a catalytic DNA hairpin. The free end of the hairpin consists of a primer, a, and the sequence to be amplified, b, as well as their complements, a* and b*. A strand-displacing polymerase extends the primer to copy b, and the extended primer, ab, then spontaneously dissociates from the hairpin while being replaced by the original b domain. Another primer can then bind to the hairpin, extend b, and continue the amplification cycle.
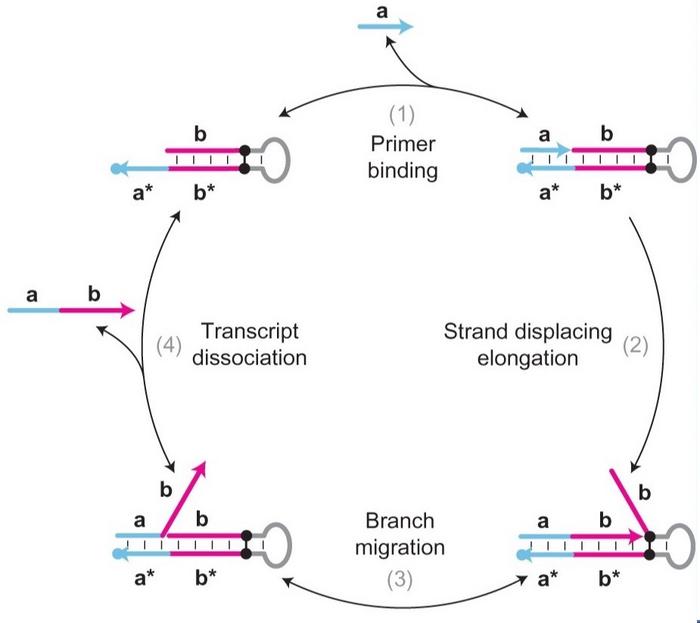
In Liu and Wan’s protocol, the primer is blocked from binding to a* by a DNA aptamer, a short oligonucleotide capable of binding to a specific target molecule. Aptamers form complex secondary and tertiary structures and bind to their target through various non-covalent interactions, such as hydrophobic or electrostatic interactions and induced fitting. They are both produced and designed entirely in vitro, in the latter case most often through in vitro evolution.
Activated Cas12a initiates the electrochemical signal
The aptamer used in this study was specific for E. coli O157:H7. It is also complementary to a* and will bind to the hairpin and block access by the primer in the absence of the target. However, when the pathogen is present, the aptamer will dissociate from the hairpin and bind to the bacteria, thereby giving the primer access to bind its complement a*. Dissociation of the aptamer will therefore initiate the PER amplification cycle and lead to continuous generation of the ab ssDNA molecule.
To show proof of concept, the authors ran a series of initial experiments without including the aptamer so that the reactions could run freely. First, they used polyacrylamide gel electrophoresis to demonstrate that a PER product of the expected length was produced only when all the necessary reagents - hairpin, primer, dNTPs and polymerase - were present.
Then the authors introduced CRISPR-Cas12a to act as a first step in the detection of the PER product. First, a sgRNA was designed to match the single-stranded PER product, and when this was specifically cleaved by the nuclease, the nuclease’s collateral trans-cleavage activity was unleashed. The presence of the PER product will, in other words, cause Cas12a to cleave any present ssDNA unspecifically.
As the second step in the detection process, an unspecific target was introduced in the form of the hairpin structure of a molecular beacon (MB) attached to a gold electrode by only one of the strands in its stem. Unspecific Cas12a activity will cleave the single-stranded hairpin of the MB and cause the unattached stem to dissociate, thereby leaving only the attached stem. A series of polyacrylamide gel electrophoresis analyses showed that, as expected, the single-stranded hairpin of the MB was cleaved only in the presence of the PER product.
E. coli O157:H7 is detected quantitatively in a broad range
The final third step in the detection process involved the gold electrode. As the MBs are gradually cleaved, the electrode’s electrochemical conductivity will change and decrease the charge transfer resistance (Rct). This change can be measured by differential pulse voltammetry (DPV) as a decrease in the current (A, ampere).
The researchers subsequently added the aptamer and a surplus of the target E. coli O157:H7 together with Cas12a and sgRNA to test the assay's functionality. As expected, this caused the DPV readout to be flat, indicating that the MBs were cleaved, so the current remained low. If any of the reagents were missing - including the bacterial target - the CRISPR-Cas12a cleavage process could not take place, and the MBs would stay on the electrode and lead to a peak in the current signal. As expected, this was also observed.
To further validate the assay, the authors added different concentrations of E. coli O157:H7. They observed a gradual decrease in the current in response to an increase in bacteria from 10 to 108 CFU·mL-1. The decrease was linear in the range of 10-106 CFU·mL-1, indicating a broad quantitative detection range. Moreover, the detection limit was 19 CFU·mL-1 based on a signal-to-noise ratio of 3. These results are superior or at least comparable to other previous aptamer or antibody-based electrochemical detection assays for E. coli O157:H7 and other pathogenic bacteria.
Liu and Wan and their co-workers also tested the detection system in diluted milk samples spiked with different concentrations of the target bacteria. In this real-world condition, the relative ratio of measured to actual concentration ranged from 97% to 116% in the range from 103-106 CFU·mL-1. However, at a bacterial concentration of 107 CFU·mL-1, the ratio was more skewed at 128%.
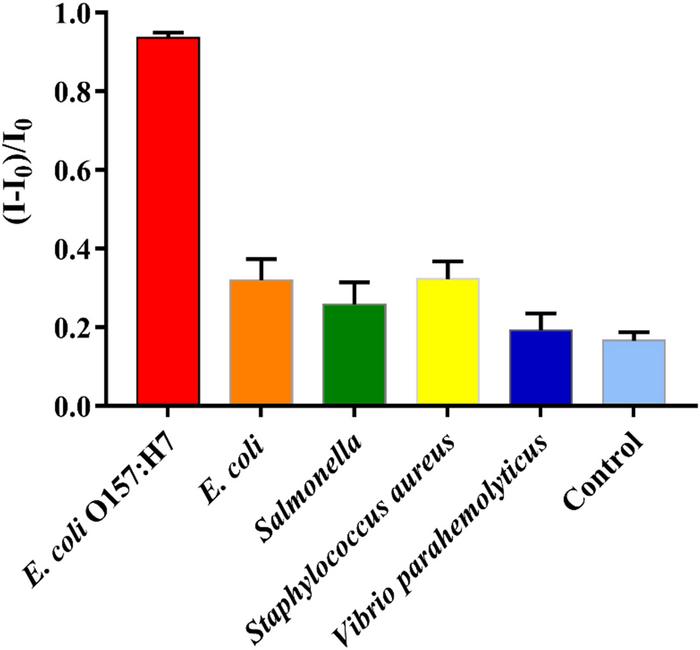
Finally, the specificity of the assay was tested using other bacterial strains and species. These experiments showed a markedly higher decrease in current - indicated as an increase in relative change of current - for the target, E. coli O157:H7, compared to common E. coli, Listeria monocytogenes, Salmonella typhimurium, and Vibrio parahaemolyticus.
The authors note that this is the first time CRISPR-Cas12a has been combined with PER to detect E. coli O157:H7 without nucleic acid extraction. They conclude that the new method could provide a rapid and effective electrochemical platform for point-of-care detection of E. coli O157:H7 that is especially relevant in resource-limited environments.
Link to the original article in Sensors and Actuators B: Chemical:
Tags
CLINICAL TRIALS
Sponsors:
Wave Life Sciences Ltd.