Strategies to Avoid and Reduce Off-Target Effects
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
All right, all right, all right, we are back!
We know, you have spent the last week on the edge, wondering what the heck is wrong with us. First, we sell you the marvels of CRISPR, just to tell you right after that this technology can misfire and have serious consequences. And then we leave you with that cliffhanger. What’s going on here?
First off, take a deep breath.
All great technologies must still be handled with care. Besides, it wouldn’t be exciting – and fun – if nature did not throw at us a curve ball every now and then.
However, scientists thrive on challenges. And in today’s chapter, we’re going to talk about some of the strategies they have developed to make CRISPR/Cas precise and ultimately safer!
So if the last chapter was a kind of “The Phantom Menace”, with off-target effects challenging the worth of CRISPR/Cas, let today’s episode be “ A New Hope”.
Modulating CRISPR staying in the cells, or how to not overstay your welcome
Let’s start with an easy, yet effective strategy.
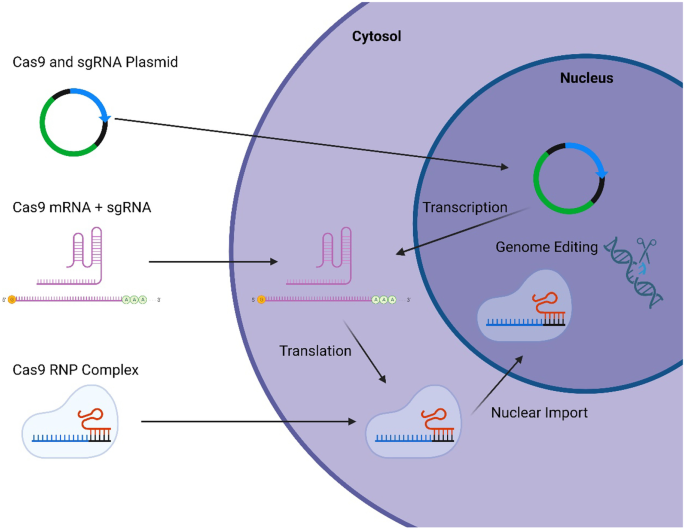
We already know that CRISPR/Cas is fairly efficient at introducing a double-strand break (DSB) in a matter of hours if not less. Now what you maybe don’t know is that, in most CRISPR/Experiments, both Cas9 and the sgRNA are delivered as a circular, double-stranded DNA molecule called a plasmid. This is the standard technique when you want to express an artificial genetic element or transgene because plasmid DNA is cheap to produce and easy to use…and it can linger for several days in the cells.
Here is the catch, however, when we use plasmids for gene editing: imagine what it means when the plasmid meant to express CRISPR/Cas stays in your cells for days. Since CRISPR only needs a few hours to do its job, all that extra time gives it ample opportunities to get itself into trouble by finding and cutting other sequences elsewhere in the genome, hence introducing off-targets. Think about it like babysitting your friend’s puppy. You can leave it alone in your living room for ten minutes and it will play with its ball as you told it to; but go away for a few hours and you may find half of your couch is missing!
For this reason, keeping Cas9 inside our cells for only as long as it’s needed is a great way to avoid off-targets. To achieve this, instead of delivering Cas9 and the sgRNA as a DNA plasmid, we can deliver them as an RNA molecule. The advantage of RNA is that, after being used, it is usually degraded by the cells within 48 hours. This way, CRISPR/Cas stays long enough to cut the on-target but not as long as if it was delivered as plasmid DNA, thus reducing off-targets. So, we just give the puppy back to our friend before it can get itself in trouble!
This can be improved even more by delivering the Cas nuclease directly as a synthesized protein, which is gone in less than 24 hours.
Nowadays, CRISPR delivery as a protein has become standard among labs focusing on human applications, and a number of providers like Synthego and IDT now offer among their products Cas9 proteins ready to transfect.
So that’s it? Problem solved, just like that?
If you are asking this, and if you’re starting to know us, you already know that the answer is far from being yes. While this is a good mitigation strategy, there is still much better we can do.
So let’s continue and step up the level a little bit!
CRISPR-nickases: it takes two
Scientists can be very creative problem solvers, and what comes is a great example. First of all, let’s remind ourselves that Cas9 nuclease has two functional domains involved in the cleavage of the DNA, called HNH and RuvC. The HNH cleaves the DNA strand whose sequence is complementary to the sgRNA, while the RuvC cuts the opposite strand. As you may imagine, mutating these domains can remove their DNA-cutting ability. Indeed, Cas9 versions with both domains mutated known as dead Cas9, which only bind to the DNA but don’t cut it at all, have been produced and used for other purposes, which we will discuss in the future.
But what happens if we mutate only one of the two domains? In that case, we will have a Cas9 which can only cut one DNA strand. Therefore, contrary to a wild-type Cas9, this mutant can only nick the DNA, and because of this it is referred to as nickase or nCas9.
And how does this reduce the off-targets? Since a nickase only cuts one DNA strand, to generate a DSB with a nickase we will need to have two nicks (one at each strand) at a specific distance, and they will need to happen very close in time. In practice, this means we will design two sgRNAs which will bring two nCas9 molecules on two different positions of the same target, and if they nick the DNA at the same time, then we will have two nicks which will amount to a DSB. This more stringent requirement will make it more difficult for off-targets to appear because the CRISPR system would need to recognize the off-target sequence not once, but twice! If only one of the two sgRNAs recognizes an off-target, the single-stranded nick that is produced will just be repaired, and no DSB will appear.
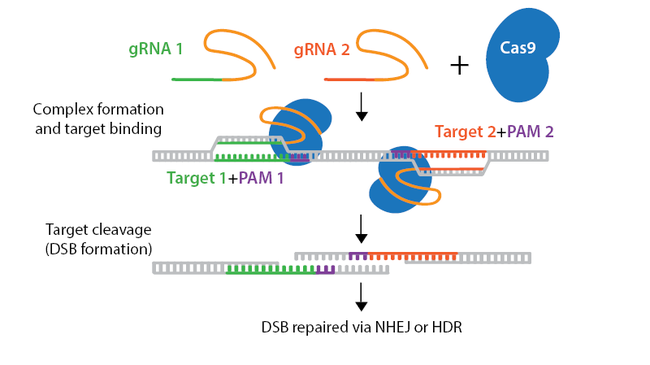
The chances for two nickases sharing the same-off target site and cutting it are much lower compared to using standard CRISPR and one sgRNA. And, in fact, scientists were pleased to observe that the rate of off-targets compared to wild-type Cas9 was much lower!
High-fidelity CRISPR/Cas: making a sharp knife safer
By now a few of you will wondering how convenient it can be to use CRISPR/Cas if you should avoid using it as a plasmid and you must go through the trouble of mutating it into a nickase and using two sgRNAs to make two nicks which turn into a single DSB.
We probably got close to a stroke by just writing this sentence.
Luckily for you – and our blood vessels – scientists did not leave it there, and they decided to give the wild-type Cas9 one more chance. How did they do that? We are so glad you asked.
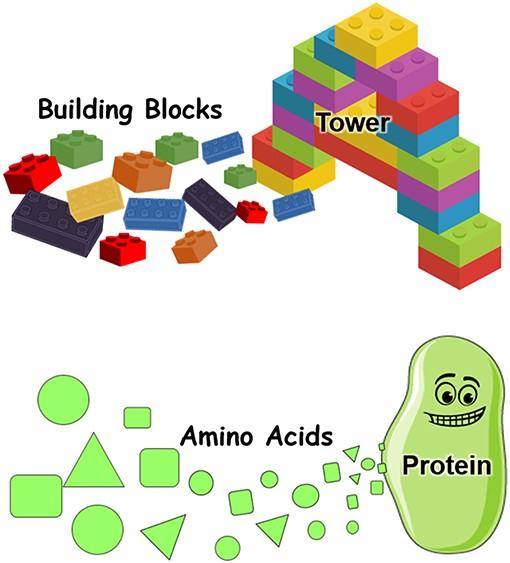
By looking at the Cas9 protein itself and identifying the key portions which make it recognize and interact with the DNA, we can engineer Cas9 to be more accurate with its selection. But how do we engineer a protein? A protein is built of a series of aminoacids, which are kind of like LEGO blocks. And we can substitute one or more of those LEGO blocks with other ones that are similar but have different connections or shapes, which will affect how Cas9 interacts with the DNA.
This can, for example, reduce the binding affinity of the Cas9 protein to the DNA. Imagine that you have the ON-target – which is a perfect match for the Cas9 – and an OFF-target which is not quite a perfect match. This engineered Cas9 will have some trouble binding the ON-target due to its reduced binding affinity, but it will be even more troublesome for it to bind – and cut – the OFF-target!
However, this nice theory takes quite an effort to bring to reality if we want the Cas9 to maintain its elevated on-target activity while reducing its off-targets. The trick is knowing which LEGO blocks to replace, and what to replace them with!
When it comes to generating more precise versions of Cas9, which we will call high-fidelity variants, there are two main things you can do: you can take a rational approach and change determined areas of the protein or you can “evolve” a new Cas9 by random mutagenesis.
Rational Mutagenesis: This approach was used in the first attempts at generating a safer Cas9. Scientists like Ben Kleinstiver looked at the structure of the Cas9 protein to understand how it interacts with the target. By modifying a key amino acid of the DNA-binding domain, they could weaken the interaction between Cas9 and the DNA so that the conditions for the Cas9 to bind and cleave it would be harder. From this, the two first safer Cas9 nucleases were born, called eSpCas9 and Cas9-HF1.
Now, rational design is not always easy and could be limited by how much you really know about the function of the different domains.
Random Mutagenesis: With this method, where we let Mother Nature do the job for us. In this approach, it is possible to “evolve” a new Cas9 by introducing random mutations in its sequence. So we’re basically switching LEGO blocks at random and seeing what effect that has. Of course, if we have a thousand or more mutants, we won’t be able to test them all one by one. So how do we know which work – and so are worth testing – and which don’t?
Scientists have developed cleaver screening approaches where a randomly mutated Cas9 is given to bacteria or yeast together with an sgRNA and two targets: the on-target and the off-target. These assays allow them to isolate - based on a “life/ death” or “ white/ red” out-put - Cas9 that are more precise while maintaining an elevated ON-target activity.
This type of screening gave birth to one of the most-used Cas9 variants of today which is the High-Fidelity (HiFi)-Cas9, developed by the group of Mark Behlke; as well as others like the SNIPER-Cas9 from Jin-Soo Kim’s lab and evoCas9 from the lab of Anna Cereseto.
One important advantage of high-fidelity variants is that they can be safely used as a plasmid without the need to further mutate them into nickases.
We hope that you will agree with us that there is still light at the end of the tunnel.
Still, designing a safe CRISPR approach is a combination of multiple factors. In the past, we discussed how a proper sgRNA design can make a huge difference. Combining this with high-fidelity Cas9 variants and proper risk assessment via off-target analysis is the key to making CRISPR/Cas a viable technology, especially for clinical applications.
So, is that the end?
Well, if you are a Star Wars fan, you know the Empire always Strikes Back, so in the next episode we will tell you how CRISPR/Cas may still raise some worries even when it only cuts the on-target site.
But don’t worry, our heroes of the CRISPR rebel alliance have already been planning for it!
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







