An Arms Race Can Make the CRISPR Toolkit Even Better
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Approximately half of all studied bacteria and 90% of archaea use CRISPR-Cas systems to defend themselves from phages by recognising and eliminating hostile mobile genetic elements. Although this defence is effective and can ensure bacterial survival, it is a complex piece of machinery involving many genes and proteins, and it comes with a substantial metabolic burden.
To reduce the costs, prokaryotes have evolved multiple mechanisms to control CRISPR-Cas gene expression so the defence can be shut down or activated according to needs. On the other hand, phages hit back and try to counteract the defence via a diverse set of anti-CRISPR proteins (Acrs) that inhibit CRISPR-Cas activity and thus help the phage to succeed in attacking bacteria.
»Mechanistically understanding how CRISPR is regulated can reveal the strengths and weaknesses of CRISPR-Cas systems. This could help us to precisely control the timing and location of CRISPR-Cas function and improve CRISPR as a genome-editing tool,« says Haridha Shivram.
CRISPR-Cas Systems Are Highly Regulated
Shivram earned his PhD in molecular biology and bioinformatics at The University of Texas at Austin before starting as a postdoc in Jennifer Doudna’s lab at the University of California, Berkeley in 2019. His current research focus is to discover novel regulators of CRISPR-Cas systems, and he is the first author of a recent review in Nature Chemical Biology that discusses the mechanisms of natural CRISPR–Cas regulatory biomolecules and the potential applications of these regulators.
CRISPR-Cas systems are divided into two classes, six types and 19 subtypes based on whether foreign nucleic acids are degraded by a single large Cas-protein or by a complex of multiple proteins, and on unique, signature proteins used almost exclusively in that subtype. For example, the well-known Cas9-system belongs to class II since it depends on a single, large nuclease. The Cas9-containing type II CRISPR-Cas system can be further divided into three subsystems (A-C) depending on their employment of the ring-shaped DNA-binding protein Csn2 or the endonuclease Cas4.
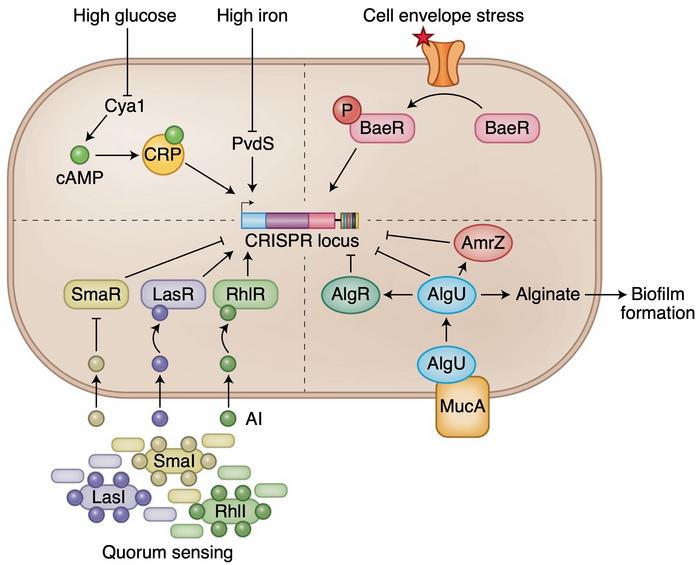
A number of transcription factors and RNA regulatory mechanisms confer transcriptional and post-transcriptional control over the CRISPR-Cas system in each of these types and subtypes. The presence or absence of viral infection can up or down regulate the defence mechanism, and bacteria can also adapt their expression of CRISPR genes to external stimuli. Through a cascade of regulatory proteins, limiting glucose and iron levels can promote CRISPR-Cas gene expression and thus adapt expression to metabolic stress induced by phage infection.
CRISPR Writes the History of a Cell Into Its DNA
»The activity and existence of CRISPR-Cas systems could vary based on the environment, nutrient availability and the likelihood of bacteriophage onslaught. In the absence of infection, it is likely that the CRISPR-Cas system may not be critical for survival and the host might shut off CRISPR activity,« says Shivram.
“Depending on the experiment and the intended application, these regulatory mechanisms are most useful for controlling or limiting CRISPR activity for a specific time and location”Haridha Shivram
Quorum sensing, biofilm formation and cell envelope stress can also adjust the defence system according to needs. The risk of phage infection depends heavily on cell density and growth lifestyle, and in such vulnerable conditions, CRISPR-Cas gene expression is increased to prepare for an attack and strengthen immunity. Shivram believes that researchers could exploit some of these regulators in their experimental or clinical work.
»Depending on the experiment and the intended application, these regulatory mechanisms are most useful for controlling or limiting CRISPR activity for a specific time and location,« he says. One obvious use of such regulatory mechanisms in therapeutical applications would be to ensure tissue, cell, and even organelle-specific editing activity and minimise off-target editing.
Moreover, the predictable control of gene editing to spatiotemporally dynamic cellular cues will enable development of molecular recorders. Such a recorder can write the history of a cell’s exposure to a given signal over its lifetime and make the recording retrievable by sequencing. This approach has already been used to track exposure to small, exogenous molecules and to record an inflammatory response over time.
Phages Fight Back With Anti-CRISPR Proteins
So-called anti-CRISPR proteins (Acrs) are another means to control CRISPR-Cas activity. Acrs are highly diverse proteins produced by phages, and as their name implies, they inhibit CRISPR activity by a variety of mechanisms and to different strengths and specificities. A majority of the known Acrs are competitive inhibitors with a negatively-charged surface that binds to the nucleic acid binding site on the Cas effector. Other Acrs are allosteric inhibitors that trap the Cas effectors in inactive conformations. Moreover, enzymatic inhibitors can modify the Cas effectors and thus inactivate multiple Cas complexes simultaneously.
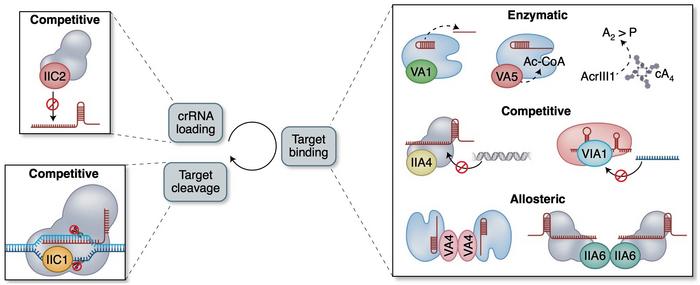
»Anti-CRISPR proteins are a major example of the adaptations and counter adaptations that affect the CRISPR war. Phages have evolved these proteins that specifically inhibit CRISPR-Cas systems from their host bacterial species. It is possible that in order to escape these anti-CRISPR proteins, bacteria, in turn, have evolved variants of CRISPR-Cas systems,« explains Shivram.
Acrs have been exploited experimentally to reduce off-target editing by reducing the time available for Cas nuclease activity and increase tissue-specific editing by controlling their presence in various tissues. Moreover, Acrs have been used to interrupt gene drives and thus to avoid unwanted ecological consequences.
Regulation Opens up for Countless Possibilities
Shivram believes that engineered regulation of these control mechanisms will be the best way to exploit them in the lab or clinic. He says:
»Domains fused to the effector proteins directly or to their transcriptional components would allow us to control the intensity of CRISPR activity and its biological location.«
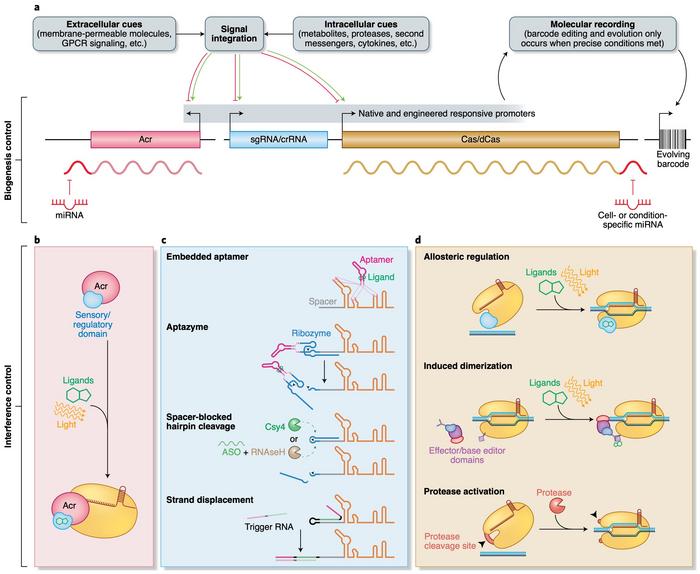
This approach is already being exploited with dead Cas9 fused to base editors and transcriptional regulators, and similar methods are also used in natural systems. For example, as the review points out, an accessory protein, RspWYL1, is believed to bind to Cas13d and function as a sponge that attracts multiple ssRNAs for cleavage by the nuclease.
“The most fascinating aspect for me is how the arms race between the host and their invading mobile genetic elements can lead to such unique molecular innovations that can both help enhance immunity but also reveal their hidden vulnerabilities”Haridha Shivram
Other approaches include coupling Acrs to light- or ligand-activated domains for tight spatiotemporal control of Cas-nuclease activity or fusing Cas-nucleases to effector proteins that can be regulated by ligands or light. The easily programmable and flexible nature of CRISPR and the insight into how phages have evolved to avoid it open up for countless possibilities for further development of CRISPR-based experimental and clinical research.
»The most fascinating aspect for me is how the arms race between the host and their invading mobile genetic elements can lead to such unique molecular innovations that can both help enhance immunity but also reveal their hidden vulnerabilities. These evolutionarily driven strategies can help us to make the CRISPR toolkit even better,« Shivram concludes.
Link to the original article in Nature Chemical Biology: https://doi.org/10.1038/s41589-020-00700-7
Tags
ArticleInterviewNewsOff-targetanticrispr proteinsBase editorsCRISPR-Cas
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







