Anti-CRISPR proteins: A smart way to make gene therapy safer

The evolutionary arms race between bacteria and their viruses, bacteriophages, have been raging for eons. In the struggle of adaptations and counter-adaptations, bacteria evolved the CRISPR-Cas immune system, which scientists have harnessed as a powerful tool for genome-editing. Now a new study shows how to use the bacteriophage counter-measure called anti-CRISPR proteins to optimize the genome editing tool.
»The way I think of it is that the bacterial CRISPR immune system provides the tools for genome editing, and the phages provide a regulatory layer to control these tools,« says the researcher behind the study, Dominik Niopek at the Heidelberg University Hospital, Germany.
The anti-CRISPR proteins suppress the CRISPR immune response, and by using them to manipulate the enzyme kinetics Niopek and colleagues reduce the off-target editing while maintaining the on-target editing.
»We can easily improve the specificity by factors between 2 and 10 fold,« says Dominik Niopek.
Constraining the CRISPR-Cas
Anti-CRISPR proteins have attracted a lot of interest since 2016, when the first was found, that are specific to the most commonly used genome editing CRISPR-Cas systems such as Cas9.
»We asked ourselves how can we harness these anti-CRISPR proteins to get better control over the CRISPR-Cas genome editing?,« says Dominik Niopek.
In 2017 a study by the Jeniffer Doudna's lab showed how timing could improve the specificity of Cas9.
The Cas9 endonuclease has difficulties, so to speak in discriminating between very similar sequences in the genome and if given enough time, all possible on- and off-target sites will be saturated and cut. However, the Doudna-lab showed that cutting with CRISPR-Cas9 for some time and then adding anti-CRISPR proteins resulted in very efficient on-target editing and much less efficient off-target editing.
»So there was basically a gain in target specificity,« says Dominik Niopek.
»And the reason for this is different kinetics behind the editing of an on-target locus and an off-target locus.«
By constraining time, one can favor the optimal enzyme substrate over the not-so-optimal substrate.
It is what the researchers call a principle of kinetic insulation, and the study gave Niopek and colleagues the idea to use weakened anti-CRISPR proteins as 'insulators' rather than constraining time.
Dominik Niopek
Dominik Niopek is a group leader of the Synthetic Biology Group in the Health Data Science Unit at Heidelberg University Hospital, and his research focuses on how to control CRISPR-Cas genome editing better. They have also developed switches to turn on and off genome editing. One example is an optogenetic switch based on engineered, light-sensitive anti-CRISPR proteins that give control of genome editing, and may be developed for gene therapy applications in the eye.
Finding the sweet spot
To explore the idea rationally, they first developed a mathematical model using enzyme kinetics to describe the basic rules of the genome editing process. The model explains well why confining CRISPR-Cas activity will separate the on- and off-target activity provided there is a difference in affinity of the Cas9 guide RNA complex to the on- and off-target sites.
They use attenuated versions of an anti-CRISPR protein called AcrIIA4 that target S. pyogenes Cas9, and show how to tune the conditions to maximize specificity.
»We can hit the 'sweet spot', so to say in the kinetic profile of genome editing,« says Dominik Niopek.
It will never be 100% on-target and 0% off-target, but with a strong affinity difference, the concept can achieve a ten-fold or higher increase in specificity, according to Niopek.
Regulatory layer on top
Importantly, where most other approaches to improve gene editing are focusing directly on Cas9, the anti-CRISPR protein strategy is like a regulatory layer that can be applied on top genome editing tools like base editors and prime editing and make them even more precise and safer.
»One can now combine these different control modalities to achieve a much more specific and confined activity profile of CRISPR Cas9,« says Dominik Niopek.
»I think the strongest point is that we are rationalizing in quantitative terms, the kinetic insulation of on- and off-target editing events. Thereby, we can predict how much one has to reduce the activity of the Cas enzyme to efficiently insulate on- and off-target editing events.«
A catch-em-all tool
As a next step, the researchers are now looking at broad-spectrum anti-CRISPR proteins that can target more than one Cas ortholog.
»I think this provides a regulatory mechanism, which is outstanding. If we can use these broad-spectrum anti-CRISPR protein to implement this strategy, then this will be a means to control, the whole plethora of Cas orthologs out there with just one kind of tool,« says Dominik Niopek.
Anti-CRISPR proteins
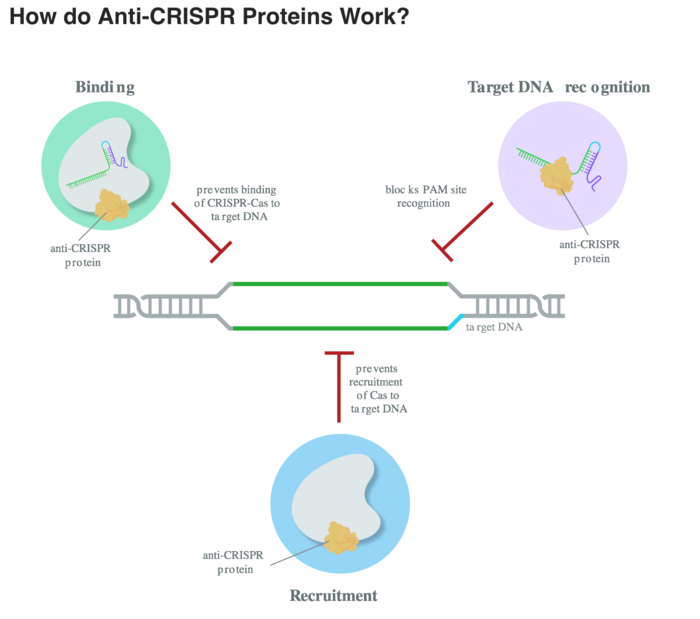
Anti-CRISPR proteins (Acr) were discovered in 2013 as phage-encoded genes that suppress the bacteria CRISPR immune response and allow phages to survive. The first Acr were specific to the type I CRISPR system, and since 2016 researchers have also characterized Acrs for the type II class such as Cas9, which are most commonly used in genome editing. Now more than 50 Acr has been characterized with different means of blocking the CRISPR system - some block the Cas endonuclease by mimicking the DNA-target, some inhibit the catalytic function, and others have enzymatic activity and cleave guide RNAs bound to Cas. (See figure above, courtesy of Synthego, who made this nice article on antiCRISPR proteins on their blog The Bench).
One example is AcrIIA4, which was one of the first Acr targeting S. pyogenes Cas9. It is a very potent Acr that combines two mechanisms of inhibition: It mimics the PAM sequence and blocks the catalytic function.
Tags
ArticleInterviewOff-targetQuality Controlanticrispr proteins
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







