Base Editing, or How to Edit the DNA Without Cutting It
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Welcome back to Making the Cut! In the last two weeks, we discussed what off-targets are and why they are important to consider when designing a CRISPR strategy. Furthermore, we covered techniques and technologies we can use to measure off-targets and avoid them. We know, it was a lot of information! But we hope you enjoyed it!
So far, every technology we talked about at Making the Cut has, indeed, worked by cutting the DNA causing a DNA Double-Strand-Break (DSB) . However, in this week’s episode, we’re going to talk about an alternative gene editing approach that, while it uses CRISPR/Cas9, no longer requires cutting the DNA to edit it. This technology, called Base Editing, has been a big revolution in the gene editing field, and we’re here to tell you all about it! So strap on, because it’s going to be a fun one!
Why cutting the DNA is not always a good idea
We know that you may be a bit confused right now. We have spent the last 8 episodes hyping up CRISPR and telling you how to use it to cut the DNA, and now we tell you it’s not a good idea? Bear with us for a second, we promise it will make sense.
While cutting the DNA is a valid option to knock out a gene, or to insert an external DNA sequence, this methodology requires us to damage the DNA of our cells. Luckily, our cells are pretty good at repairing that damage, which we sometimes exploit in our advantage, as discussed in Part 2. However, these repair mechanisms can fail, leading to a DNA lesion that may be repaired wrongly, or not repaired at all. And as we discussed in Part 6, the cell has some surveillance mechanisms that can recognize when that happens. And they’re normally not very happy about it!
As genomics trailblazer George Church once said “ genetic engineering sometimes is an act of genome vandalism“.
Indeed, these mechanisms can decide that the DNA damage is too severe and that the best idea is to kill the cell. If you think about it, it makes complete sense: our cells are part of a larger organism, and it is better for some damaged cells to commit the ultimate sacrifice than potentially risk damaging the whole organism. “And how would they damage the organism?”, we hear you ask.
Severe DNA damage could unbalance the delicate equilibrium that controls cell growth and division, leading those damaged cells to uncontrollably grow and reproduce, thus forming a tumour. Indeed, other types of DNA damage, for example that caused by solar UV radiation or exposure to carcinogenic substances, are the main drivers of our cells turning into cancerous ones.
So what about we don’t give our cells any bad ideas?
That’s exactly what some CRISPR scientists thought about. And they decided that the best way to avoid this would be to do gene editing without cutting the DNA. And no, they were not dreaming.
Base editing can edit the DNA without cutting it
So far, we have been focusing a lot on the DNA-cutting capabilities of the Cas9 protein. However, in Part 3 we also mentioned its other amazing ability: using an RNA template to find DNA sequences and bind to them.
This ability is going to be essential for the approach that we will discuss, so if you haven’t read Part 3 already, what are you waiting for? We’ll wait patiently for you to be done.
All done? Good. Now think about what would happen if we removed Cas9’s ability to cut the DNA. As we mentioned in Part 8, we can mutate the two DNA-cutting domains, called HNH and RuvC, thus turning a radar-guided scalpel, into a docile cotton bud. But what would happen to Cas9 then? Well, it would still be able to find the DNA sequence as instructed by its sgRNA, but not able to cut it, so it would just hang around there. Kind of like when you go to your best friend’s place without any particular plan, just to hang out with them. That’s what we call a dead Cas9 (dCas9).
And that’s where the brilliant idea comes in. The scientists in David Liu’s lab at Harvard University decided to bind the dCas9 to another enzyme called a deaminase. This is a very interesting type of enzyme which can chemically modify nucleotides. Without going into much detail, we will give you the main gist to better visualize how it is possible to modify nucleotides using a deaminase.
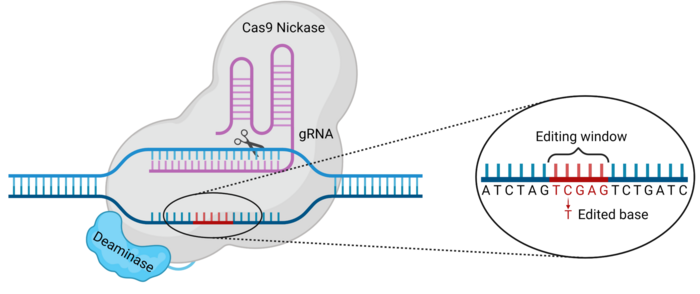
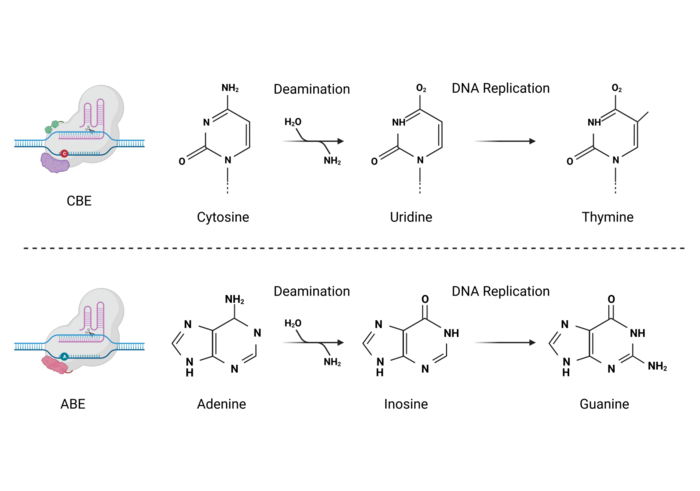
As you know, our DNA is composed of 4 nucleotides called Adenine (A), Guanine (G), Thymine (T), and Cytosine (C). Now these are not the only existing nucleotides. For example, in our RNA, there is no thymine, and instead, we can find another nucleotide called Uridine (U). What’s interesting about these nucleotides is that their structures are quite similar. For example, Uridine looks pretty much like Cytosine. The only difference between them is that Cytosine has an amino group whereas Uridine has an O2 group.
Now, remember we said we were going to use a deaminase? What do you think this enzyme would be able to do? Yes, you guessed right, you brilliant geek! A deaminase removes amino groups.
Here’s the bonus points question: What would happen if this deaminase found a Cytosine nucleotide and removed its amino group? If you said that Cytosine will be converted to Uridine, you deserve a cookie!
Okay, so by binding a deaminase to dCas9, we can bring this complex, which we will call a base editor, to a specific DNA site. The deaminase can recognize a Cytosine nucleotide within the target sequence and convert it into Uridine. But what effect will that have?
If you remember, we said before that Uridine replaces Thymine in the RNA. And our cells know that very well. In fact, they know it so well, that when they find a Uridine nucleotide in the DNA sequence, they don’t really freak out. They’re more like “oh, somebody put a Uridine here instead of a Thymine, typical… It’s fine, I’ll fix it.”
Do you realize what just happened? We tricked our cell! By deaminating Cytosine into Uridine, we made the cell think that there was a Thymine there, and now there is one! So we’ve effectively replaced a Cytosine with a Thymine, modifying the genetic sequence! That’s what we will call a C-to-T edit.
Go ahead and tell us that’s not awesome, we dare you!
Different base editing tools can introduce various edits
Okay, now that you have an idea of how base editing works, let’s discuss what we can actually do with it. As we said, the first base editing experiments substituted a Cytosine with a Thymine. That’s because the deaminase they bound to dCas9 was a Cytosine deaminase, which means that it can only deaminate cytosine. But what would happen if we replaced that Cytosine deaminase with another enzyme? As you might have guessed, we would be able to introduce other types of edits.
The second type of base editors that came out used an Adenine deaminase. Interestingly, when we deaminase Adenine, it turns into another nucleotide called Inosine. And while Inosine is not in our DNA or RNA, it is also something that our cell recognizes frequently. Interestingly, Inosine can be found on our DNA when a Guanine nucleotide has been damaged. So, again, we’re tricking our cell. We’re making it think it has to repair a damaged Guanine nucleotide, when it was an Adenine all along!
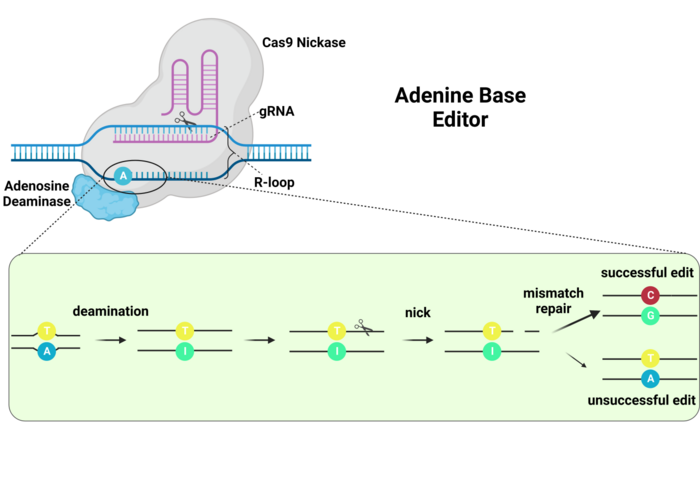
So that’s two types of edits we can make, C-to-T and A-to-G. It doesn’t seem like a lot, but what’s interesting is that more than 60% of the disease-causing mutations we can find in humans are either G-to-A or T-to-C. That means that with only these two types of base editors we could already potentially cure a great majority of genetic diseases that are caused by point mutations. Not bad, is it?
In addition to that, some other types of base editors have been developed to target rare types of mutations. This way, C-to-G edits, as well as simultaneous C and A edits are now also possible.
Base editing has limitations
Okay, so far we’ve told you how cool this new base editing technology is. But this wouldn’t be us if there wasn’t a twist somewhere along the line. So, what’s the matter now?
Luckily, this is not as dramatic as in previous episodes. Base editing is indeed an awesome technology. However, some limitations have been found in the last years. But worry not, because this is the bread and butter of research scientists!
Bystander Effect: The first and most important limitation of base editors is the bystander effect. That may sound similar to the off-target effects we talked about recently, and you would be right. In this case, however, the effect is not far away in the genome, but very close. In the same target sequence, in fact.
Because what we told you about above was a perfect example, where the deaminase finds and edits a single Cytosine nucleotide in the target sequence. But what happens if there are two Cytosines there? Or more? Well that’s where bystander effects come into play.
Most times, we’re going to want only one of the Cytosine to be edited, but the deaminase is not that smart, and it may also modify the other Cytosines around it, resulting in a sequence we did not want. To avoid this, new base editors have been developed that have smaller “Editing windows”, that is the range of nucleotides that the deaminase can see.
Edit size: The other main limitation of base editing is that it can only be used to introduce single-nucleotide edits. While those are extremely important and have many applications, other gene editing strategies require substituting several nucleotides, or even inserting large DNA sequences at a target site. For those strategies, base editing cannot be useful.
Off-target effects: The last limitation we will discuss are off-target effects. Yes, base editors can also recognize the wrong sequence and edit it. While the damage will, in most cases, not be as bad as with regular CRISPR, the wrong edit at the wrong location could still have dramatic effects, so better to reduce those! And interestingly, base editors have raised another issue: since deaminases are not great at differentiating DNA and RNA, base editors can also have off-target effects in the RNA. We’re not going to go into detail on this point just yet, but this was kind of a big deal and new editors have reduced deaminase activity in the RNA, which has partially solved this issue.
As you are reading this, we are sure we are all vibrating on the same line “ this technology is awesome, can’t wait to see what the future holds!”
Luckily for you, this is not Stranger Things, and you will need to wait only one week to read all about it.
In fact, next week, we will take a step back from the technical side and discuss CRISPR therapies that are close to approval. Don’t miss it, it’s going to be a great one!
Meanwhile, if you’re left wanting more on how base editing works, CRISPR Medicine News published a more extensive and technical Explainer on Base Editing, where you will find your curiosity satisfied!
**CRISPRMED25 - The 2nd CRISPR Medicine Conference, Copenhagen, Denmark, April 7-11, 2025**
Learn about the latest discoveries in base editing at the CRISPRMED25 Conference in Copenhagen, Denmark, April 7-11, 2025.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







