Breaking: Click Editing Takes on Prime Editing for Precisely Writing Genomes
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
“We sought to develop a genome editing approach that enables the facile and precise installation of nearly any genetic change into a chromosome while also overcoming challenges of other technologies”Benjamin Kleinstiver
Genome editing technologies have revolutionised molecular biology, enabling precise modifications to the genetic code. Traditional methods, such as CRISPR-Cas9, have been widely adopted for their ability to introduce targeted changes with relative ease. However, these techniques often come with limitations, including off-target effects and the need for double-strand breaks (DSB), which can lead to undesirable mutations, chromosome alterations and cell cycle arrest.
Recent advancements have sought to address these challenges, aiming for higher precision and reduced collateral damage. Among these innovations, base editing and prime editing have proved successful but still carry limitations in, for example, the scope of possible edits, efficiency and ease of use.
"We sought to develop a genome editing approach that enables the facile and precise installation of nearly any genetic change into a chromosome while also overcoming challenges of other technologies," says Benjamin Kleinstiver, who led the study. He is an assistant professor at Massachusetts General Hospital and Harvard Medical School and senior author of a paper that was published today in Nature Biotechnology.
To do this, his team - including the paper's co-first authors Joana Ferreira da Silva and Connor J. Tou - developed a sophisticated system of coupled enzymes. The click editor (CE) is a fusion protein that incorporates an RNA-programmable DNA nickase, a DNA-dependent DNA polymerase (DDP), and a single-stranded DNA (ssDNA) tethering domain derived from HUH endonucleases (see Figure 1).
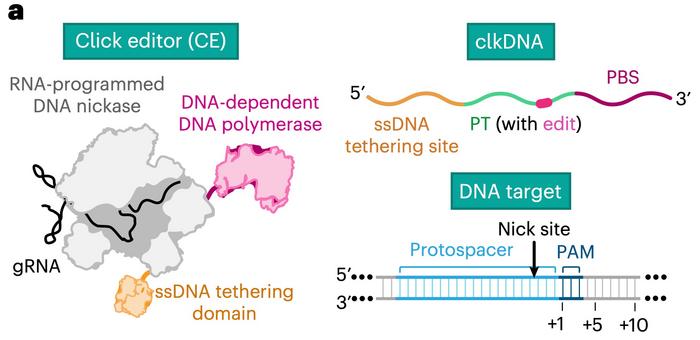
This combination allows the CE to target specific genomic loci guided by RNA. The HUH endonuclease domain covalently attaches "click DNA" (clkDNA) templates, which encode the desired edits, to the genomic DNA at the nicked site. The DNA polymerase then uses these templates to incorporate the genetic changes precisely.
"Our study describes the development of a first-generation click editor, which uses wild-type enzymes and minimally or unmodified nucleic acid templates called clkDNAs," explains Benjamin Kleinstiver.
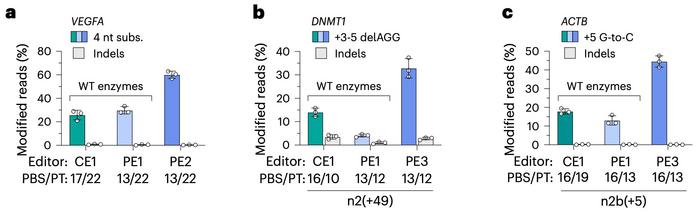
Click editing demonstrated lower off-target effects compared to traditional CRISPR-Cas9 nuclease approaches, similar to prime editing. The new method achieved precise genome modifications such as substitutions, insertions, and deletions with efficiencies of up to 30% in various human cell types, including primary fibroblasts. In some instances, click editing achieved higher efficiencies than PE1 (prime editing version 1) but generally lower efficiencies than PE2 (prime editing version 2) and PE3 (prime editing version 3), which use engineered reverse transcriptase domains for enhanced performance (see Figure 2).
**CRISPRMED25 - The 2nd CRISPR Medicine Conference, Copenhagen, Denmark, April 8-11, 2025**
Learn about the latest discoveries in CRISPR Medicine at the CRISPRMED25 Conference in Copenhagen, Denmark, April 8-11, 2025.
Notably, the modular design of click editing allows for rapid and high-throughput screening of clkDNA templates with various modifications, providing flexibility in optimising editing conditions. This scalability is an advantage for fine-tuning the method to achieve higher efficiencies and specific editing needs.
"We are optimistic about future engineering approaches to improve both the click editor protein and clkDNA templates, and we envision this will improve click editing efficiency and safety. Furthermore, we are excited about expanded conceptions of click editing that may include additional editing types and lengths, including those for precise and safe exon- or gene-sized sequence installation," says Benjamin Kleinstiver.
The study was led by researchers from Massachusetts General Hospital and Harvard Medical School, including Joana Ferreira da Silva, Connor Tou, and Benjamin Kleinstiver. The findings were published today in Nature Biotechnology. A preprint of the study was published last year on the bioRxiv server. CRISPR Medicine News has priviously covered click editing and its competitors in this article.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.