CRISPR-Associated Transposases - Precise Integration of Large Gene-Editing Cargos Without DNA Repair Requirements
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
In nature, CRISPR-Cas functions as an ancient defense system against foreign invaders, such as bacteriophages, transposable elements and plasmid DNA. Recent studies suggest that these invaders may have found ways to not only evade CRISPR-based defense systems, but they may even exploit them for their own benefit.
The CRISPR-associated transposases (CASTs) are one good example of a strategy used by a foreign invader to exploit CRISPR-Cas systems. CASTs were discovered when two research groups found that either the CRISPR-Cas subtypes I-F or the CRISPR-Cas subtypes V-K were found within the same loci as Tn7-like transposons in some bacterial genomes (see Fact Box). The I-F and V-K subtypes were found in Vibrio cholera and cyanobacteria, respectively, and were named CRISPR-associated transposases because of their similarity to Tn7-like transposons.
The Tn7 transposon
The Tn7 transposon is a mobile genetic element (MGE) found in many prokaryotes such as Escherichia coli (E. coli). By using a self-encoded transposase enzyme, a transposon can duplicate itself and move within the genome. The transposon co-exists with its host, for whom its propagation is usually unharmful, which allows the transposon to maximise its diffusion.
The Tn7 transposon encodes five genes: TnsA, TnsB, TnsC, TnsD, and TnsE. In particular, TnsA and TnsB – with the support of TnsC – are required to excise the DNA sequence of the transposon from one region of the host genome, after which it will be inserted into another site in the genome. The TnsD and TnsE proteins interact with the so-called TnsABC core machinery, and Tn7 preferentially directs insertions into conjugable plasmids when TnsABC interacts with TnsE. When TnsD interacts with TnsABC, Tn7 preferentially inserts itself downstream into the bacterial chromosome.
CASTs might look like CRISPR-Cas but they are really just hijackers
Several features of the I-F and V-K CRISPR-Cas subtypes have stood out during their discovery and characterisation so far.
Firstly, none of these Cas proteins possess nuclease activity. The I-F subtypes are characterised by a multiple Cas protein complex (Cascade) which lacks the Cas3 nuclease component that is known to degrade viral DNA as part of CRISPR-based immunity, while the V-K subtypes contain only the Cas12k effector whose nuclease activity is naturally inactivated. So, unlike the well-known CRISPR-Cas9 system that is used in labs all around the world, CASTs are unable to introduce a double-stranded break (DSB).
Secondly, Tn7-like genes are located upstream of Cas-encoding genes, and finally, the presence of tnsA (missing in CRISPR systems V-K), tnsB, tnsC, and tniQ genes, which are some of the key components of the Tn7 transposon, hinted that the CASTs may possess transposon-like activity.
Although tnsB, tnsC and tniQ are present within CASTs, the tnsE gene, which is needed by Tn7-like transposons to promote transposition in certain DNA regions, is missing. This led to the hypothesis that CASTs were hijackers, where the essential role of TnsE could have been taken over by CRISPR-Cas, and the gRNA may be exploited for transposition in a target-specific fashion.
Early characterisation of CASTs
The fact that transposons can harness the CRISPR-Cas system to facilitate their propagation presents an attractive new approach to targeted integration of DNA into genomes.
Seeing this huge potential, Dr. Feng Zhang’s lab at The Broad Institute investigated the CRISPR-Cas subtype V-K, which is mainly found in cyanobacterial species. Zhang’s team chose the V-K subtype because it requires only one Cas effector protein, Cas12k, making it much simpler to work with than a multi-protein complex such as the I-F CRISPR-Cas subtype.
Within two species of Cyanobacteria (Cytonema hofmannii and Anabaena cylindrical), Zhang´s team identified spacer sequences that matched known cyanobacterial plasmids, thus substantiating the hypothesis that CASTs may have mediated transposition into other mobile genetic elements (MGE). After characterising the essential element of the V-K subtype (the tracRNA, crRNA sequence and the PAM), the team then tested the possibility of targeted insertion by transposition.
DNA repair-independent targeted integration
When it comes to targeted integration via standard CRISPR-Cas systems, a DNA donor is supplied together with the Cas/gRNA complex. Normally, the DNA donor comprises the desired sequence harboured within two stretches of nucleotides that complement the DNA ends flanking the DSB generated by CRISPR-Cas, and these stretches are known as homology arms (HAs).
The HAs are used by the cell’s homology-directed DNA repair (HDR) pathway to identify the DNA sequence to be used as a blueprint to repair the DSB. If after the DSB, the HDR is engaged and the donor DNA is used, the desired sequence will be integrated at the target site (Figure 1).
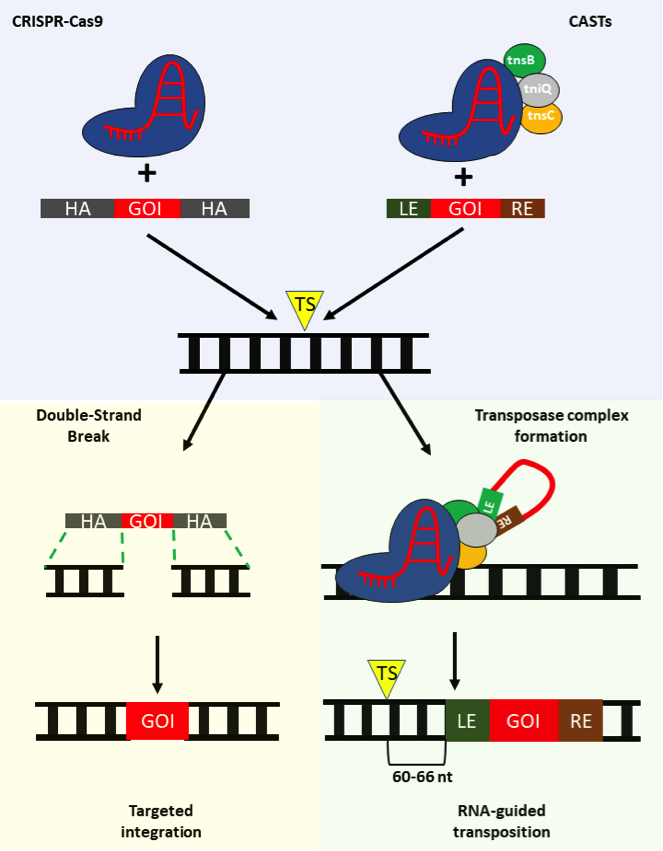
However, since CASTs do not cut DNA, they rely on a different mechanism to integrate, which is attractively independent of DSB repair mechanisms.
To perform targeted integration with a CAST, two main elements are required: i) a plasmid expressing the Tn7-like genes, the Cas12k and the gRNA, and ii) the DNA donor. In this case the desired sequence doesn´t possess HAs but it is embedded between two sequences; the transposon left end (LE) and right end (RE). In the case of CASTs, the Cas12k/gRNA will bring the Tn7-like genes close to the target site. TniQ may mediate the interaction between the Cas12k and the donor DNA. The latter would be then excised and inserted 60 to 66 bp downstream of the PAM (GTN) by the transposase complex formed by tnsB, tnsC and tniQ.
Upon delivering the Scytonema hofmannii CAST (ShCAST) to E. coli, Zhang’s team observed up to 60% directional integration events in 29 of 48 sites tested. Intriguingly, they were able to integrate up to 10 kb sequence without compromising the efficiency compared to their standard donor (2.5 kb).
However, it is important to note that off-target analysis revealed that only 50% of the integration events occurred at the desired locus, while the others were scattered in the genome. These findings indicate that, as of yet, CASTs have significant off-target activity.
Potential to address unmet needs within therapeutic genome editing
The possibility to manipulate genomes by applying small nucleotide changes or inserting entirely new sequences has already opened doors to new therapeutic applications.
The standard approach with CRISPR-Cas editing requires engagement of the HDR mechanism in order to integrate the desired DNA modification within the genome. However, HDR frequencies are usually low and almost null in post-mitotic cells such as neurons or liver cells. One possible way around this may lie in the development of HDR-independent platforms such as base editors and prime editing. However, base editors can only change one base at a time and the current maximum editing capacity of prime editors is less than 100 bp, leaving a significant limitation in terms of cargo size.
The desired approach should not be strictly dependent on HDR but should enable the possibility to integrate large modifications at the desired location with clinically relevant frequencies, and CASTs may be the answer.
Promising but still early days
Although there is every reason to be excited about CASTs, there are some key challenges to be solved before we can expect to see them in the clinic.
CASTs have only been tested in prokaryotes so far, and the technology will need to be optimised for usage in eukaryotes, particularly in mammalian cells.
In addition, in order to be integrated the donor sequence must be embedded with the transposon LE and RE sequences which will be also incorporated in the target site. This means that in contrast to HDR-dependent approaches or base editing and prime editing, genome editing with CASTs is not “scarless”. This also means that targeted integration within an exon is not suitable as the reading frame would be altered by the presence of the LE and RE sequences. Conversely, integration of large cargo in locations where the reading frame is not a concern such as intronic sequence or within the so-called safe harbour loci (see Fact Box) are an attractive prospective. Finally, further optimisation is needed to reduce the CAST off-target integration rate, which at the moment is not negligible.
Safe harbour loci
A safe harbour locus is defined as a genomic location that does not play a functional role in cell and organism biology. As such, genetic alteration within such loci won’t lead to consequent alteration of the cell physiology. One of the most well known safe-harbour loci in humans is the AAVSI locus.
To conclude, CASTs represent a novel CRISPR-Cas based genome-editing technology that facilitate integrations of large cargos in a DNA-repair independent manner. Although it’s early days yet, companies, e.g, Metagenomi are already working to find new and better CASTs to empower future therapeutically-relevant applications.
Antonio Carusillo, PhD, currently employed at Vector BioPharma.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







