CRISPR–Cas-Armed Phages Could Provide a Solution to Antibiotic Resistance, New Study Suggests

Although the advent of antibiotics in the late 1920s has revolutionised medicine, effective treatment of infectious diseases using antibiotics has become increasingly challenging because of the emergence of antibiotic resistance. In addition, the role of the microbiome in human health and the detrimental effects of antibiotics on the microbiome are becoming increasingly evident, raising additional concerns regarding the use of antibiotics.
In a recent study, a team of researchers at SNIPR BIOME used the CRISPR-Cas system to “arm” engineered bacteriophages (viruses that infect bacterial cells) to make them attack certain E. coli strains that cause infections in immunosuppressed individuals. These CRISPR-armed phages successfully reduced the E. coli burden in mice.
This study provides proof-of-concept evidence that engineered phages can overcome the limitations of traditional antibiotic treatments and fill the unmet need for alternative treatments to combat bacterial infections, particularly those caused by antibiotic-resistant E. coli strains.
»Our vision is to provide a safe and effective solution for the treatment of bacterial infections in vulnerable patients who are already suffering through side effects of cancer medication,« said Yilmaz Emre Gencay, DVM, PhD, senior scientist at SNIPR Biome and first author of the study.
»Our findings suggest that engineered phages may represent a treatment alternative to avoid additional complications resulting from the burden of antimicrobial use for the management of E. coli infections, regardless of the presence of antimicrobial resistance,« he added.
The study was led by Milan Zdravkovic, MD, PhD and Morten Otto Alexander Sommer, PhD and was published in Nature Biotechnology.
Using CRISPR-Cas to engineer phages as antibacterial agents
To develop a phage-based therapy against a diverse range of clinically-relevant E. coli, the team tested a library of 162 wild-type lytic phages on a panel of phylogenetically diverse E. coli strains to identify the phages with broad and complementary target strain coverage (Figure 1). This state-of-the-art phage screening assay led to the identification of eight phages that were specific for various E. coli strains, exhibited complementary binding to bacterial surface receptors, and were able to stably carry inserted cargo.
Commenting on the rationale for using phages as antibacterial agents, one of the study authors, Eric van der Helm, who is also VP of Scientific Affairs, Bioinformatics & Automation at SNIPR Biome, said: »Phages have evolved in an arms-race manner with their bacterial targets for billions of years, which makes them highly specialised viruses for finding a susceptible host and infecting bacteria to replicate. Ultimately, this makes them one of the best candidates for bacterial therapeutics based on gene delivery.«
To further improve the ability of the phages to target clinically-relevant E. coli strains,the team subjected them to tail fibre engineering. »We used the generated data to improve the wild-type phages by introducing an add-on receptor-binding protein to enhance their receptor portfolio,« Dr Gencay explained.
Subsequently, the selected phages were armed with the CRISPR-Cas system containing sequences specific for E. coli. Genetic engineering of phages involved homologous recombination, which led to a ~7 kb deletion in the genome of the phages while inserting the CRISPR-Cas circuit.
»A challenge was to ensure that the deleted region did not drastically affect phage performance. Therefore, sections of a large genomic region were systematically tested to ensure dual killing via both phage replication and CRISPR-Cas activity,« Dr Gencay noted. All these steps were repeated in large-scale host-range assays to compare and ensure a broad and complementary coverage over diverse E. coli strains.
The engineered phages were tested in vitro for their ability to inhibit bacterial growth. In addition, the team assessed the in vivo growth inhibitory effect of the phages on E. coli in a mouse gut colonisation model by treating mice with the engineered phages and measuring the E. coli burden. The most effective and complementary CRISPR–Cas-armed phages were selected for further development — the team termed this proprietary cocktail of engineered phages “SNIPR001”.
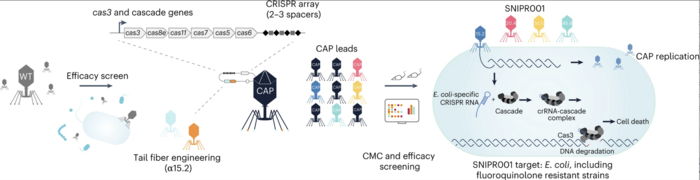
Figure 1. Schematic overview of SNIPR001 development. Wild-type phages were screened against a panel of 429 phylogenetically diverse E. coli strains. Phages with broad activity against E. coli were subjected to tail fibre engineering and were armed with CRISPR-Cas systems containing E. coli-specific sequences. CRISPR–Cas-armed phages were tested for host range, manufacturing and control, and in vivo efficacy. Gencay et al. Nature Biotechnology (2023). https://doi.org/10.1038/s41587-023-01759-y
Engineered phages target bacteria in biofilms and reduce the emergence of phage-tolerant E. coli
To minimise the risk of treatment resistance, the team designed CRISPR arrays targeting multiple regions in the E. coli genome, including virulence genes and essential genes. Co-cultures of E. coli cells and CRISPR–Cas-armed phages and their ancestral phages (1-to-9 ratio) showed a higher relative abundance for engineered phages (68%–86%) compared to wild-type phages (7%–10%), demonstrating the competitive superiority of the CRISPR–Cas-armed phages.
The ability of CRISPR–Cas to eliminate E. coli was tested in a biofilm assay. This analysis showed that the CRISPR-Cas in engineered phages successfully suppressed bacteria in biofilms, as well as reduced the bacterial counts to numbers below the limit of detection (200 CFU mL−1) compared to an empty vector upon conjugation (Figure 2).
»This study is one of the largest host-range datasets displaying various phage-host interactions with an abbreviated yet comprehensive phylogenetic E. coli diversity. Phage-host interactions often form nestedness, in which a phage with a broad host range can infect strains of another phage with less breadth. Despite this, being able to achieve a 90% coverage for E. coli strains for our indication was surprising,« Dr Gencay said.
The high killing efficiency of CRISPR–Cas-armed phages was confirmed under restricted bacterial growth conditions, which more closely reflect the in vivo conditions in the gut.
The four most effective and complementary CRISPR–Cas-armed phages (α15.2, α20.4, α48.4, and α51.5) were selected for further analysis as SNIPR001 drug product.
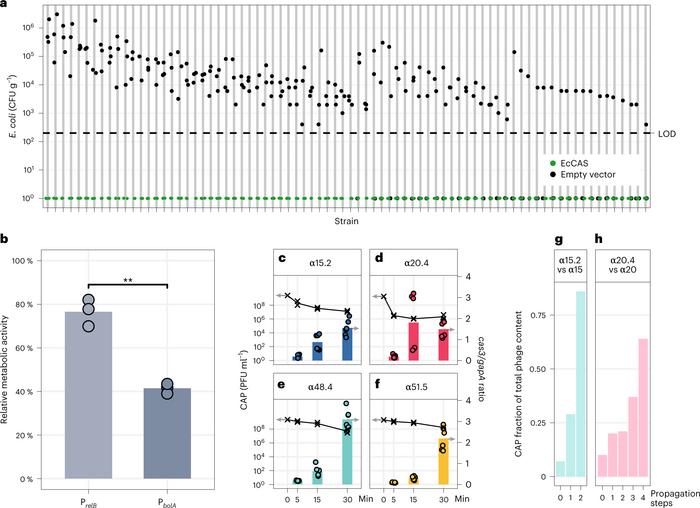
Figure 2. CRISPR–Cas can eliminate E. coli in biofilms, and engineered phages robustly express the circuit and outcompete ancestral phages. (A) CRISPR–Cas-driven elimination of E. coli clinical isolates by conjugation of CGV-EcCAS (green) or empty vector (grey). (B) Metabolic activity reduction in biofilms. (C–F) Increasing levels of Cas3 relative to housekeeping gene (gapA). (G, H) Relative abundance of engineered phages (α15.2, α20.4) and wild-type phages (α15, α20) during co-culture with susceptible E. coli strain. Gencay et al. Nature Biotechnology (2023). https://doi.org/10.1038/s41587-023-01759-y
SNIPR001 is well tolerated and reduces E. coli load in vivo
The tolerability of SNIPR001 was studied in female Göttingen minipigs. CRISPR–Cas-armed phages were detected in the faeces for up to seven days after SNIPR001 administration. However, no CRISPR–Cas-armed phages were found in the plasma, indicating that there was no systemic exposure to SNIPR001. No clinical signs of adverse events were observed.
The efficacy of SNIPR001 in reducing the E. coli burden in vivo was evaluated using a mouse gut colonisation model, in which mice were administered streptomycin to reduce bacteria in the gastrointestinal tract and then inoculated with E. coli. SNIPR001 treatment began two days after inoculation and lasted for two days, with three daily doses administered.
Phages successfully passed through the gastrointestinal tract and led to a dose-dependent reduction in E. coli colonisation, with the high dose resulting in a significant reduction of 4 log10 CFU g-1. The combination of the four CRISPR–Cas-armed phages (i.e., SNIPR001 cocktail) was more effective than any single phage in reducing colonisation, and no major resistance to the engineered phages was observed in the tested bacteria.
Challenges and further clinical development
Recently, SNIPR Biome reported positive interim results from an ongoing Phase 1 clinical trial. Overall, the data suggest that oral dosing of SNIPR001 for seven days across three dose levels is well tolerated, and adverse events are limited to mild to moderate side effects. The interim results also suggest that SNIPR001 treatment leads to presence of CRISPR–Cas-armed phages in the gastrointestinal gut and reduces the E. coli burden in the gut.
»Our recently released top-line data demonstrate the feasibility of using CRISPR–Cas-armed phages in a clinical setting and provide proof of principle that engineered phages could provide an alternative for the treatment of antibiotic-resistant infections,« said Milan Zdravkovic, MD, PhD, Chief Medical Officer at SNIPR Biome and senior author of the study.
The company is currently seeking funding to test SNIPR001 in Phase 2 trials. However, Dr Zdravkovic noted that securing funding has proved to be the greatest challenge in developing this method further. He explained that the business model for funding research on new antibiotics needs to be improved, as the revenues are insufficient in comparison to the investment required to bring a new antibacterial treatment on the market.
»I think that, unfortunately, people underestimate how much modern medicine actually rests on the fact that we can successfully treat even very simple infections and how much we need new antibacterial treatments for at-risk populations. Regulatory and government bodies need to find ways to better incentivise pharmaceutical companies to stay in this space,« he added.
Initiatives are also needed to address regulatory considerations for using SNIPR001 in clinical settings, as there are currently no approved CRISPR-armed phage products on the market.
Link to the original article in Nature Biotechnology:
Engineered phage with antibacterial CRISPR–Cas selectively reduce E. coli burden in mice
Christos Evangelou, PhD, is a freelance medical writer and science communications consultant.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University