CRISPR-Cas13d Nuclease System Effectively Inhibits HIV-1 Replication In Vitro

HIV is a retrovirus, which is capable of inserting copies of its single-stranded RNA genome into the DNA of an infected host’s cells, by using a virally-encoded reverse transcriptase enzyme. The virus predominantly attacks CD4+ T cells, the depletion of which can lead to acquired immunodeficiency syndrome (AIDS) and associated complications such as pneumocystis pneumonia.
UNAIDS estimates that 38 million people have been infected with HIV globally as of 2020.
Research in the last decade has greatly expanded the scientific knowledge about CRISPR-Cas systems. In more recent years, type VI CRISPR-Cas systems, which include the Cas13 family, have garnered interest in the research community, and in 2020 Cas13a was demonstrated to inhibit HIV-1 infection in vitro.
More recently, additional effector proteins within the type VI-D subtype of Cas nucleases have also been identified, including CasRx, some of which have been explored in infectious disease research with promising results.
A recent publication in Viruses has detailed a CRISPR-based strategy to effectively target and control HIV-1 infections in vitro with CasRx. The lab of Dr. Smita Kulkarni at Texas Biomedical Research Institute successfully combined a novel set of guide RNAs (gRNAs), known as polyHIV, with CasRx to target and inhibit HIV-1 replication. This work demonstrates for the first time the potential of the CRISPR-Cas13d nuclease system to target acute and latent HIV infections that could potentially be used to enhance the efficacy of current HIV treatment options.
»We are excited about CasRx and its potential to work as a new molecular scissors to search and destroy viral RNA,« Kulkarni says. “There are still many questions we need to answer especially in larger model systems, but these first results are very encouraging.«
Connecting CRISPR to HIV
Intriguing discoveries in science are often the answers to a series of questions, and this has been the case for Dr. Smita Kulkarni and her team. Kulkarni developed her interest in HIV research starting as a postdoc fellow at the National Cancer Institute, where she investigated the role of host genetic factors that affect HIV outcomes, specifically the expression of non-coding RNAs integrated in the host genome.
Current treatments for HIV include combination therapy, which relies on the use of multiple highly active antiretroviral drugs (HAART) to reduce HIV-1 viral load and maintain high CD4+ T-cell counts. The drawback of HAART therapy is its life-long commitment, since interruptions to treatment can lead to the development of drug-resistant strains.
When Kulkarni started her own group at Texas Biomedical Research Institute in 2016, CRISPR naturally found its way into her research interests:
»While we were looking at host genetic factors, we were also looking at expression of these non-coding RNAs that are in host genome, and how their expression affects HIV outcomes. And one of the better ways to control expression or regulate expression of these non-coding RNAs, for our experiment, turned out to be CRISPR!« Kulkarni comments.
Customising CRISPR to target HIV
Since HIV is a highly mutable virus owing to the inherent error-prone nature of reverse transcriptase, Kulkarni’s team strategically chose four sites in the HIV genome that are more than 70% conserved among HIV-1 transcript sequences deposited in the LANL database, and HIV-specific gRNAs were designed against these sites, as Kulkarni explains:
»Targeting HIV RNA is not so simple, because HIV is a highly mutable virus,« Kulkarni explains, who continues: »To address this, we decided to look at certain structural features of HIV-1 RNA that are conserved between isolates, taking into account that not every region of RNA can be easily targeted by CRISPR because of tertiary structure formation.«
With these criteria in mind, four gRNAs were chosen out of 20 candidates to form a poly-gRNA assembly (polyHIV) and paired with CasRx, a derivative Cas protein of the Cas13d family with high RNA cleavage activity. The chosen target sites were non-overlapping, and were located in the regions encoding gag, pol, protease (prot), integrase (int), cPPT, and central termination sequence (CTS) in the HIV-1 genome. The study is the first to deliver a single vector with four gRNAS targeting non-overlapping conserved regions of HIV-1.
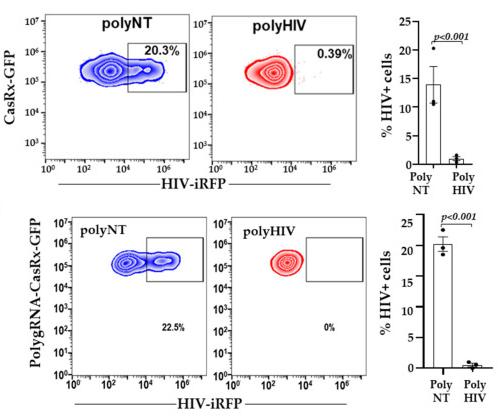
The novel CasRx system was capable of inhibiting HIV-1 production in lentivirally-transfected cells of human embryonic kidney origin (annotated as ‘LRx cells’) in as little as 48 hours (Figure 1).
In this experiment, LRx cells were transfected with plasmids encoding the CasRx protein combined with multiple gRNAs (polyHIV) or a non-targeting control gRNA (polyNT) along with a molecular clone of HIV (HIV-iRFP) encoding a near infrared fluorescent marker (iRFP-670). Flow cytometry was used to measure HIV-1 replication within the LRx cells, as indicated by a percentage change in iRFP-expressing cells (% HIV+ cells).
In LRx cells transfected with polyHIV, HIV-1 replication was reduced by more than 90% compared to polyNT control cells, as indicated by both the percentage of iRFP-expressing cells and production of infectious HIV particles (Figure 1 top). Similarly, when a lentiviral construct with all 4 gRNAs and the CasRX-GFP was used, there was an even higher inhibition of HIV+ cells (Figure 1 bottom).
These experiments were also repeated in primary CD4+ T cells, with CasRx cloned into a mammalian expression vector and transfected into active CD4+ T cells along with the same 4 gRNAs (polyHIV). When HIV-iRFP virus particles were measured, T cells transfected with polyHIV expressed significantly fewer virus particles compared to the polyNT control, providing hopes that the CasRx approach may also work in vivo.
Efficient targeting of HIV-1 reservoirs
Patients diagnosed with HIV often harbour viral reservoirs, which exist as proviral HIV-1 DNA that is integrated in the genomes of infected resting memory CD4+ T cells at tissue sites that include the brain, lymph nodes, blood, and digestive tract. These reservoirs allow the virus to remain in an inactive state and evade immune clearance in a phenomenon known as latency.
To address whether their approach could address HIV-1 reservoirs emerging from the genomic DNA into the cytoplasm, Kulkarni’s team tested the efficacy of their polyHIV and CasRx system in degrading HIV transcripts reactivated from J1.1 cells.
J1.1 is a chronically infected latent cell line derived from HIV-infected Jurkat cells that when stimulated, can reproduce active HIV. To ensure that the CasRx was localised to the cytoplasm, they replaced the nuclear localisation signals (NLS) with the MAPK nuclear export signal (NES), resulting in CasRx-NES.
Kulkarni’s team nucleofected J1.1 cells with plasmids encoding polyHIV or polyNT and CasRX-NES, with the GFP tag serving as a tool to track luciferase expression (and indicating transfection efficacy). The transfected cells were then stimulated with a latency reversal agent for 24 hours, after which supernatant was collected and added to another cell line. This cell line contained an integrated reporter gene for firefly luciferase under the control of an HIV-1 long terminal repeat, and was exposed to the supernatant for 48 hours. This experiment revealed that the supernatant from cells transfected with polyHIV exhibited an approximately 8-fold reduction in luciferase activity compared to polyNT control cells, indicating that polyHIV together with CasRx is capable of inhibiting reactivated HIV-1 from latently-infected cells (Figure 2).

Taking advantage of CasRx
Unlike current CRISPR-based strategies used to target HIV, such as Excision Biotherapeutics’ clinical-stage Cas9-based candidate EBT-101, which is designed excise HIV proviral DNA using CRISPR-Cas9 and two gRNAs, Kulkarni’s team relied on CasRX to target HIV replication, in an approach that leaves genomic DNA intact:
»CasRx can be used as a corrective tool to specifically target the desired RNA sequence, in this case from HIV-1 RNA, and the beauty of CasRx is that it doesn’t touch DNA at all,« Kulkarni says.
Originally derived from the microbial strain Ruminococcus flavefaciens, the CasRx nuclease is an engineered Cas13d ortholog that was found by others to exhibit strong nuclease activity in human cells, and was capable of knocking down endogenous RNA and maneuvering alternative splicing for precise transcriptome targeting.
Despite its prokaryotic origins, CasRx’s activity in a broad range of species has made it an intriguing tool to implement in CRISPR-based targeting strategies. Kulkarni notes that CasRx is beneficial in hindering the replication of acute viral infections, and it has recently been used to detect SARS-CoV-2 sequences from both synthetic and patient-derived samples.
»Sequences that are highly mutable can be quickly targeted, because all you need to know is its sequence, where in the body it is, and which cells it infects. Then it is just a matter of designing guide RNAs, for which now algorithms are available,« Kulkarni says.
A supplement to other HIV therapies
Major hurdles in developing a functional cure against HIV are viral diversity and the persistence of proviral DNA despite lifelong highly active antiretroviral therapy. While CRISPR-spCas9 can eliminate latent proviral DNA, viral diversity limits the widespread efficacy of CRISPR-Cas9.
CasRx with multiple gRNAs targeting conserved sites in HIV could be used as a combinatorial regimen with latency reversal agents in ‘shock and kill’ approaches to reduce the viral burden and block new infection by the reactivated virus.
While the present work demonstrates the potential of CasRx to target HIV-1 reservoirs and the use of several gRNAs may help to impede the generation of escape variants, Kulkarni points out that CasRx may need to be combined with other drugs to make targeting HIV in humans more efficient:
»HIV may be a much more difficult target than acute viral infections. We may have to still combine it with drugs or other therapies because HIV forms deposits and causes chronic infection.«
It is also important to note that an approach based solely on Cas13 will not be sufficient to eradicate HIV, since the virus integrates into the genome of infecting cells and persists as a latent reservoir in macrophages and CD4+ T cells - an issue that has also impeded current HAART-based treatments.
Novel strategies to address these long-standing challenges are emerging, with one example being the application of base editing to disrupt the major HIV-1 co-receptors. In a recent publication in Molecular Therapy, researchers led by Mark Osborn at the University of Minnesota Medical School demonstrate that genetically disrupting CCR5 or CXCR4 HIV-1 co-receptors through base editing could serve as a useful strategy to protect human CD4+ T cells from HIV-1 infection.
Despite technological advances in targeting approaches, it is worth noting that CRISPR-based strategies to tackle HIV require more monitoring, with respect to potentially harmful by-products that result from gene editing. In a recent publication in Virology, researchers in Italy and the UK report that while CRISPR-Cas9 is successful in removing HIV-1 provirus from infected cells, extra attention must be paid to free-floating provirus that may develop after gene editing. The authors of that study caution that gene editing may increase proviral DNA circles with restored long terminal repeats (LTRs), and that this caveat must be considered when developing CRISPR-based HIV-targeting strategies in the future.
Future directions with HIV and CRISPR technology
Overall, the use of CasRx to improve the treatment landscape for HIV is at an early but exciting stage. The in vitro results published by Kulkarni’s group demonstrate that CRISPR technology is effective at targeting and suppressing HIV-1 infection, but Kulkarni acknowledges that the work needs to continue in order to demonstrate proof of concept in vivo and in clinical trials. The current study uses transfection-based delivery, which is not translatable to the clinic. To address this, Kulkarni and her team are currently investigating how to deliver their polyHIV and CasRx system into cells via lipid-based delivery systems versus using lentiviral vectors:
»We are collaborating with Southwest Research Institute, which is a sister Institute of Texas Biomedical Research Institute, to be able to put these molecules in a vector form or in a liposome encapsulated form in vivo,« Kulkarni says.
These experiments are being conducted in primary cell lines to ensure that delivery with lipids is effective, with the next steps involving primary cells from HIV patients to assess potential off-target effects. If successful, this delivery method could serve as a safer alternate to lentiviral vectors. A similar delivery approach was explored by researchers from the University of Nebraska Medical Center, who recently used lipid nanoparticles to deliver CRISPR-Cas9 systems to latently-infected CD4+ T cell and monocyte-macrophage cell lines, reporting success with blocking HIV-1 reactivation.
When asked about the future directions of HIV and CRISPR, Kulkarni had this to say, »We'll have to target HIV simultaneously at multiple sites, I think that's one take home message.«
There is still more work to be done before we can rely on CRISPR-based strategies alone to treat HIV successfully in the clinic, but the latest research from Kulkarni’s group and others seems to suggest we are headed in the right direction.
Link to the original paper published in Virology, from which Figures 1 and 2 are adapted:
Efficient Inhibition of HIV Using CRISPR/Cas13d Nuclease System.
Priya Rangan is a scientific communications specialist for a nutraceutical company in Italy and a freelancer science writer.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleInterviewNewsViralLentivirus (LV)Human Immunodeficiency Virus Infection, HIVCRISPR-CasCas13
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







