Curing One Base at a Time
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Welcome back to Making the Cut! Today is the 10th episode of this series!
If you are reading this, that means that not only have we managed to engage you in an exciting – yet quite complex – topic as genome editing, but also that you are now part of a growing community of science lovers who can better appreciate how mankind has progressed towards precision medicine!
So, a big round of applause and another cookie!
As promised, for this week’s episode we will tune down the “technicalities” knob a little bit and focus on the applicative side of Base Editing.
So, if you have not read Part 9 yet, where we explained how Base Editing works, we recommend you do so before discovering how we can cure disease, one base at a time!
Short recap
By now you know that the two main base editors are Uncle ABE (Adenosine Base Editor) and Aunt CBE (Cytidine Base Editor), which allow us to change A -> G and C->T respectively.
Last time, we told you that more than 60% of human-associated genetic disorders are associated with single-point mutations. But this can be a little vague, right? So, let’s bring up some relevant examples before we tell you about human clinical trials.
Oh yes, you read it right, Base Editing is being rolled already for human application.
Base editing corrects LMNA in a mice model of progeria, the genetic disease that makes people age faster
The first disease we will talk about is the Hutchinson–Gilford syndrome, mostly known as Progeria. This name with Greek origins says it all. It refers to premature aging.
Children affected by this condition appear healthy at birth but start developing the first sign of the disease by the second year of life. These young patients have unusually wrinkled faces, small, fragile bodies, stiff joints, and hip dislocation. The most severe symptoms include kidney failure, loss of eyesight, atherosclerosis, and other cardiovascular problems. This eventually leads to death by the 15th year of life.
The cause of this devastating disease are mostly mutations in the LMNA (laminin A) gene, which codifies for the laminin A protein, the key component of the nuclear lamina. This can be pictured as a filament scaffold that holds the architecture of the nucleus together ensuring proper DNA replication and RNA-transcription.
Mutations in the LMNA gene lead to the production of an aberrant form of laminin A that can no longer serve its function, resulting in a nucleus that appears like a bleb instead of being rounded.
Unfortunately, disrupting the faulty gene with CRISPR/Cas9 as it was done with TTR gene for example – see the previous episode – won’t help much. Cause we still need the healthy LMNA product.
This is where Base Editing comes to the rescue.
Adenine base editing can correct LMNA mutations
One of the most common LMNA mutations (>90% of all cases have this same mutation!) is a C1824T. This means that in the coding sequence of the LMNA gene, the nucleotide number 1824 is a Thymine (T) instead of being a Cytosine (C). This alters the way the mRNA is synthesized and processed, eventually leading to a truncated protein.
C to T, uh? Well, it sounds like a job for Uncle ABE!
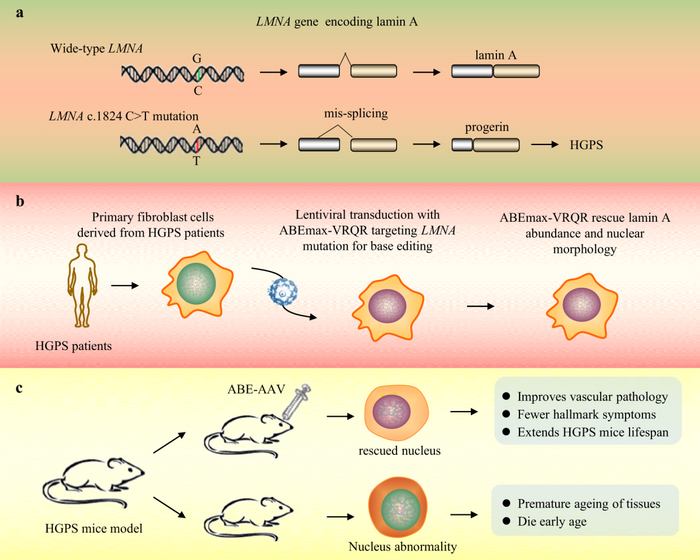
Wait, but didn’t we say last week that Uncle ABE converts A to G? You are not wrong, but you also shouldn’t forget that the DNA has two strands that are complementary to each other. This means that on the complementary strand, this C-to-T mutation is represented as a G-to-A mutation. Thus, by targeting this A at the 1824 position on the opposite strand with ABE, the A will be replaced by a G. When this happens, the DNA repair machinery – that we just tricked – will insert a C on the other side, which is our actual sequence
And voilà! The mutation has been corrected!
This way, the team led by David R. Liu could correct up to 60% of the mutated LMNA in a mouse model, prolonging its life expectancy, with some of the treated animals almost regaining a normal life expectancy! Is that cool or what?
Base editing and sickle cell disease, plan B!
You all remember for sure the great news about Casgevy, the first CRISPR-based therapeutics to be approved for use in patients – beyond clinical trials – in the UK.
This therapy could successfully treat sickle-cell anemia and beta-thalassemia, making it possible for patients to become independent from recurrent blood transfusions and other medicaments.
While this is already a great milestone, there is always room for improvement.
Base editing is a safer option to treat SCD
As we know, Casgevy uses CRISPR-Cas9 to introduce a double-strand break (DSB) to reactivate the expression of fetal hemoglobin to compensate for the mutated adult counterpart in the affected patients.
But you also know that DSBs can be quite aggressive, so scientists have kept on working on better ways to address this. In the case of Sickle-Cell-Anemia, scientists – among which David Liu once again, this guy is everywhere! – developed a brilliant strategy revolving around base editing.
This may seem easy, but it was not a trivial task.
SCD is mainly caused by a mutation in the HBB gene which modifies a small part of its sequence called a codon from GAG to GTG. So A>T and if you recall neither of ABE and CBE can actually do that.
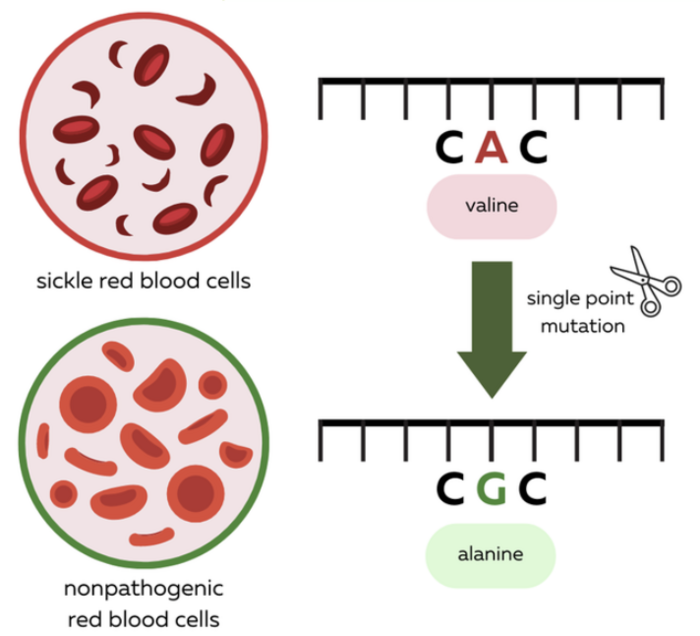
So what do you do? Well, at the time there were no other options. But scientists got very creative and clever and recalled a case report about a mutation in a patient in Indonesia whose HBB gene has neither of the two codon versions we mentioned above. This patient presented a GCG codon instead, which results in a benign mutation with no side effects whatsoever on the function of hemoglobin. This new version of hemoglobin was dubbed the Makassar hemoglobin, after the region where this was identified for the first time.
This is a bit like when your expensive iPhone charger cable is chewed into pieces by your cat, and they recommend you to buy only an original one. But you need to save money -to buy a bigger screen so you can enjoy WeDoCRISPR better- and decide to buy a standard off-brand cable just to find out that it works just as well.
That’s what happened here!
So, yes ABE and CBE cannot do T>A, but they can convert T>C all right!
Thereby, by altering the SCD-causing mutated HBB gene to the Makassar hemoglobin they could revert the SCD phenotype and rescue the symptoms in a mouse model, offering a valid alternative to Casgevy that will most certainly be on the approval lane soon enough!
Base editing and human applications: time to get real!
Enough with pre-clinical examples and let’s jump to settings where Base Editing is being tested as a potential line of therapy in humans!
High LDL cholesterol is involved in many diseases.
Verve Therapeutics and Beam Therapeutics teamed up to address patients affected by high-risk heterozygous familial hypercholesterolemia (HeFH), established atherosclerotic cardiovascular disease (ASCVD), and uncontrolled LDL-C levels (low-density lipoprotein a.k.a. the “bad cholesterol).
All these conditions, despite a different etiology – a fancy word to refer to the cause of a condition – have in common the accumulation of bad cholesterol.
The current line of treatments forces patients to regularly take medications like statin with a variety of potential side effects which often causes them to drop out of the therapy
In comparison, gene editing may offer a one-time-only treatment option.
Base editing can reduce blood LDL levels
As we said, the main cause of these diseases is the accumulation of circulating LDL cholesterol. So, scientists set themselves to reduce it by increasing the uptake of LDL into the liver, where it can be metabolized.
But how to do that? In normal conditions, the LDL Receptor (LDLR), which captures LDL from the blood into the liver, is not always expressed on the liver surface, but rather “cycled.” This way, it’s only there when needed.
The protein encoded by the PCKS9 gene is the main actor in charge of recycling LDLR by making it go to the lysosome – our garbage incinerator – for degradation.
Therefore, inhibiting the PCSK9 protein can prevent LDLR degradation and thus increase the presence of this receptor on the surface of the liver cells, increasing cholesterol uptake. This effectively reduces LDL levels in the blood and improves cardiovascular risk.
This is what Verve has set itself to do with the aid of Base Editing! Indeed, base editing can also be used to KO a gene! Yes, like CRISPR/Cas.
Gene KO using base editing
This can be done in three ways:
Disrupting the start codon: biology 101 tells us that all proteins translation start with an “ATG” codon. By changing this start codon with ABE ( A->G or T->) or CBE ( G->A), we can disrupt the translation of the gene, effectively knocking it out
Introducing a premature stop codon: Our genetic code has a way to tell when a protein’s sequence is over. That is the stop codon, which makes protein production end. The neat part is that we can alter a single nucleotide in the sequence of a gene to introduce a premature stop codon. For example, the codon CGA codifies for the Arginine amino acid. However, if we use CBE and change C->T, we will have TGA instead, which identifies a stop codon!
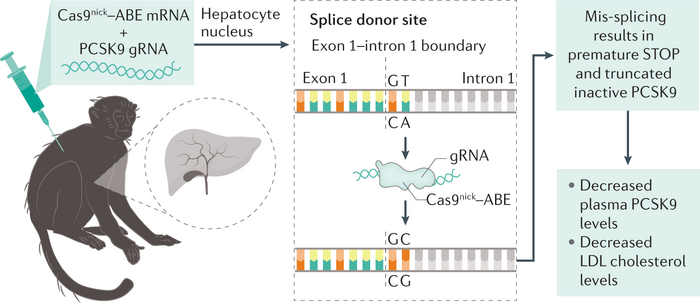
Altering Splicing: If you go back to biology101 you will remember that the mRNA splicing process is guided by a splice donor and a splice acceptor, sequences that identify the exons that need to be pasted together. Usually, they are identified by the sequence “GT” and “AG” respectively. By now your eyes are already shining, looking at how many ways all the four nucleotides could be easy prey for ABE and CBE.
By altering these nucleotides in the splice donor or acceptor, it is possible to mess up the splicing process and obtain an mRNA transcript that retains an intron, which will alter the reading frame and hopefully produce an aberrant stop codon.
This last strategy was what Verve Therapeutics used to abrogate the expression of PCSK9, up to more than 90% of PCKS9 reduction and 60% reduction in non-human primates. So far, 6 patients have been dosed with this therapy, called VERVE-101, and all are showing a tangible reduction in both their PCSK9 and LDL levels!
The next step will be to increase the cohort size to get a more comprehensive picture of the effectiveness of VERVE-101.
Base editing can generate "superhero" cancer-fighting CAR-T cells
For the second and last example of in vivo Base Editing applications , we will touch upon a topic we will talk more about in the next sessions, Chimeric Antigen Receptor (CAR) T cells.
CAR-T cells are engineered cancer bounty hunters
For the moment, picture this: T cells are engineered to recognize cancer cells with high efficiency by installing a chimeric receptor able to recognize specific protein residues – a.k.a. antigens – present on the surface of these cancer cells.
It is like giving a picture of a perp – that says “Wanted Dead or Alive” to a bounty hunter. But the “Alive” part is crossed out.
Yes, there is no “Three Strikes and You are out” with cancer cells, one felony, and down you go.
Once T cells are equipped with this chimeric receptor – we will find out later how – they can recognize and kill cancer cells with ease.
So why the need for Base Editing or genome editing at all, for that matter?
Well, the point is that generating CAR T cells is a lengthy process. The standard procedure requires isolating T cells from the affected patient, engineering them into CAR T cells, and growing them to a number able to take out the cancer cells.
This process takes time and, as you may guess, sometimes patients do not have such luxury.
So what can we do to fix that?
Making "superhero" CAR-T cells that can cure any patient
We know what you are thinking “ If only we could have CAR-T cells already stored away and save them for the dark days when a patient needs them”.
We are proud of you, as you are thinking exactly like a genome engineer.
While that’s a great idea, there are some challenges to this, particularly the compatibility between donors.
If donors are not matching – like in organ transplantation – the receiver’s immune system will recognize the CAR T as “non-self” (a.k.a. “intruders”) and attack them. This can both disrupt the therapy per se and further endanger the patient.
Lucky for us, the genetic components behind the “self/ non-self” recognition are well studied, and this is where Base Editing comes in.
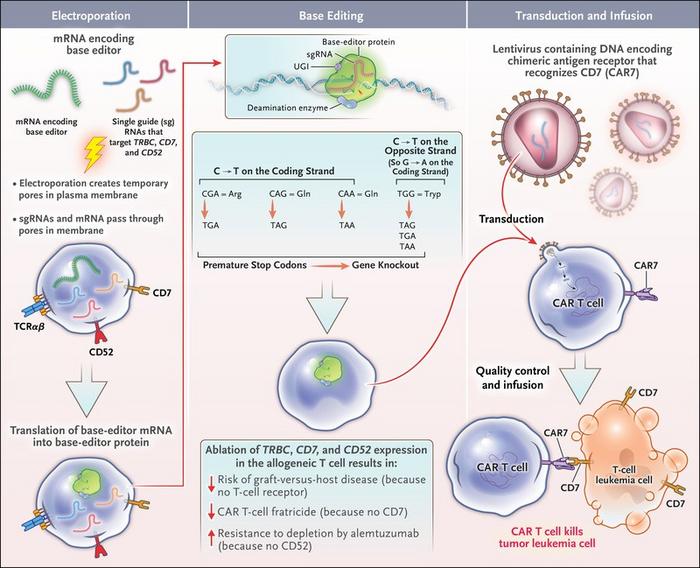
Base-edited CAR-T cells have already saved lives!
This is the story of Alyssa, the first ever treated patient with base editing. She was affected by an aggressive form of T-cell leukemia. Because of this, it was not really possible to use Alyssa’s own T cells to engineer them into CAR-Ts.
They needed healthy ones. To do so, scientists got T cells from a donor and, together with installing the CAR receptor, they blocked the expression of the genes involved in the “self/ non-self” recognition pathway, thus rendering these new CAR-T cells universal. Also, they modified an additional gene which renders the edited cells resistant to a chemotherapeutic drug used in the clinic to kill leukemic T cells. They made superhero CAR-Ts!
We are happy to say that Alyssa recovered from her leukemia, becoming a living milestone of what Base Editing is capable of. And hopefully, many more patients will benefit from this approach.
With this, we come to a conclusion – for the moment – on the topic of knocking genes off or disrupting them. As some of you may have been already wondering “breaking things off” is not all a genetic engineer can do. What happened to introducing new information or making larger modifications.
In the next issue, we will talk about another hot topic in the genome editing community: targeted DNA integration!
Stay tuned, we will see you in the next one!
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.