Designing Your First CRISPR Experiment
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
All right, let’s get right into it.
Last time we saw how to direct the nucleolytic activity of the Cas9 nuclease towards a desired genomic location, we needed to program the sequence of its guiding unit named sgRNA.
But how do we do that, practically speaking?
That’s what we are going to find out today.
So, let’s plan a CRISPR experiment!
- The goal of the experiment
First off, let’s define the goal of this experiment. To keep it simple, we will aim to lose the expression of a gene. In scientific jargon, to knock it out!
What do we need to do that?
In the second episode of this series, we have seen how when a Double-Strand Break (DSB) is introduced, the cells try to repair it quickly and, in this attempt, mistakes – known as insertion or deletions, indels – can be introduced. If this happens in the coding sequence of a gene, the way the information contained within is read - the reading frame – may be altered. This alteration may even read to aberrant stop codons, which can result in unstable RNA transcripts or truncated proteins leading to the eventual knockout (KO) of the gene .
So the first point is clear, we need to target an exon!
This brings us to last week’s article regarding the requirements to target a genetic sequence, that is the need for the presence of a consensus sequence – known as the Protospacer Adjacent Motif (PAM) – immediately next to our desired target.
We are sure you are all going like “Aha, I remember that!” and start appreciating how everything is slowly coming together.
So, we must target the coding part of the gene – an exon – and the exon needs to have a PAM sequence.
Some of you may ask "why not targeting at the start codon, right after the promoter of the gene?" While this seems a clever thing to do – and the reasoning is good – genes may have alternative start codons within their sequence that can still drive the expression of a functional or semi-functional protein.
To ensure a full KO of a gene, it is important to hit exons comprising most of the protein and, ideally, shared among all different isoforms – due to the sneaky alternative-splicing processes – that a gene may have.
2. Identifying suitable sgRNAs
The process of designing the spacer sequence of a sgRNA – which defines the target - can be done “by hand” or using open-source tools. While open-source tools are usually the way to go, let’s have a look at how you will do it by yourself to better understand the process.
For our experiment, we have decided to target the C-C chemokine receptor type 5 (CCR5) gene. Most of you have heard of it already, since the CCR5 receptor in the membrane of lymphocytes is the main entry point for HIV. For this reason, the KO of CCR5 is a very attractive target for infection-disruptive strategies like CRISPR/Cas9.
By looking at publicly available databases like NCBI or ENSEMBLE we can access the information we need: both the genomic sequence and its corresponding coding sequence. The first tells us which are the exons and which are the introns of the gene, while the second gives us information about the different transcript variants a gene may have.
In this case, CCR5 has two isoforms whose main coding portion is encoded within Exon 2. Therefore, Exon2 is the optimal region where we can direct our search for a PAM sequence.
Once a PAM is found – in the case of using SpCas9 it is going to be “NGG” – the 20 nucleotides right before the PAM will be the targetable sequence that our spacer will be complementary to.
3. Helpful tools
But what happens if more than one “NGG” – and therefore more than one target – is available? How do we choose?
Well, this is a bit complicated, because several parameters come into play. For this reason, it would be hard to evaluate on our own. Thankfully, brilliant researchers have developed tools that can help us in the decision-making process.
One of these is CHOPCHOP.
The first thing this software does is very similar to what we just did. Once a target genetic region is provided – for example CCR5 Exon2– the software looks at the DNA sequence and the coding portion of the gene. Then it scans for PAMs and based on this it designs potential sgRNAs. And it ranks them. A machine-learning system takes into account parameters like the nucleotide composition of the target, its GC content, and others, and predicts the efficiency of a given sgRNA. While such prediction cannot faithfully recreate what will happen in a cell, it is a good estimate. This can restrict hundreds of potential sgRNAs to a few dozen that have the highest chance of mediating efficient cutting of the DNA by the Cas9 nuclease.
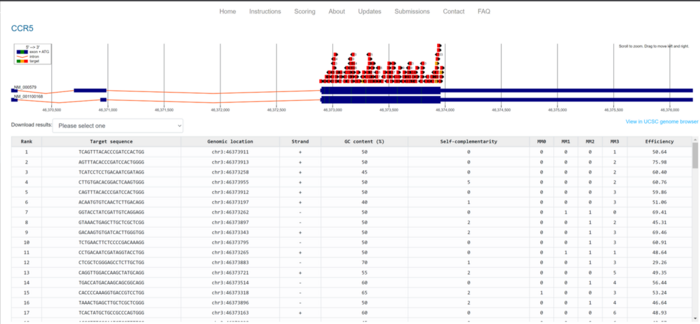
Additionally, CHOPCHOP looks at something else that is relevant: whether the sgRNA sequences that it has designed are unique to our gene. Because imagine that we selected an sgRNA that cuts our DNA very well but is also present in five other genes. We would be knocking down six different genes, and the effects would be completely unpredictable. Not to speak about the potential for severe DNA damage or even cell death, things we would probably want to avoid. Something similar could happen with sequences that were very similar to our target sequence (for example 19 nucleotides out of 20 being the same), which we call off-targets. The bigger the difference, the lower the chances Cas9 would accidentally cut an off-target sequence. Thus, CHOPCHOP compares the designed sgRNA sequences with the whole genome and finds whether each sgRNA is unique for our gene and which potential off-targets it has, allowing us not only to select the best-performing sgRNA but also the safest one.
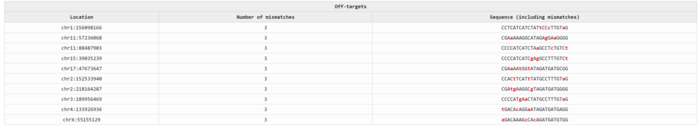
Off-targeting is a very sensible topic, and we will talk about it in the advanced stage of this series.
Tools like CHOPCHOP – combined with a proper and educated design by the experimenter – can smooth the selection of the sgRNAs to be tested for efficient KO of a gene.
4. Next episode content
Of course, after designing your CRISPR experiment and performing it, you will want to know whether or not it worked, rather than just trusting CHOPCHOP predictions or our words.
Therefore, in the next episode, we will see how we can measure the successful editing of our first attempt at knocking out a gene.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







