Epigenetic Editing Proves Link Between Epigenome Dysregulation and Neuropsychiatric Disorders
CMN WEBINAR 28st Sept. 3:00 PM CEST
Epigenomic Editing Ameliorates Detrimental Effects of Adolescent Alcohol Exposure
Register here

Increasing evidence points to the epigenome as a key influencer in human health and behaviour, and dysregulation of the epigenome has been well-documented in many neuropsychiatric disorders. However, previous studies have all been correlative; without tools to manipulate the epigenome, no direct causal links can be proven, and treatment options are limited.
Thanks to recent advances in CRISPR technology, researchers can now modify the epigenome – not only to prove causality between epigenome and phenotype, but also to provide potential therapeutic avenues (see Fact Box).
Endowed Professor in the Department of Psychiatry and Director of the Alcohol Research Centre at the University of Illinois at Chicago, Subhash Pandey has been researching the effects of adolescent binge drinking on adult alcohol use disorder and anxiety for several decades. In particular, Pandey was fascinated by early evidence that alcohol use has rapid effects on the epigenome of genes expressed in the amygdala.
Pandey’s new study, published in Science Advances, offers the first proof that epigenetic marks caused by adolescent binge drinking result in adult anxiety and alcohol use disorders in rats. Using catalytically-dead Cas9 (dCas9) fused to different epigenetic enzymes, Pandey explains that these neuropsychiatric disorders can be switched on and off without any observable off-target effects, indicating the therapeutic potential of epigenome editing (eGE):
»This study really provides some hope that this type of detrimental epigenetic alteration can be corrected, and that the phenotype can be reversed. It results in a really drastic reduction in alcohol use.«
What is the epigenome and how does epigenome editing work?
The epigenome is made up of genetic elements and chemical marks attached to DNA that control the transcription of genes. There are three key types of epigenetic modifications: DNA methylation (5mC), histone modifications, and non-coding RNA. Epigenetic chemical marks can be heritable, and they are susceptible to alteration by environmental factors.
Histone post-translational modifications include acetylation, methylation, sumoylation phosphorylation, and ubiquitylation. Because histones are responsible for packaging of DNA into chromosomes, the alteration of these proteins can impact gene expression. The main focus of the present study is histone acetylation, which is regulated by histone acetyltransferase and deacetylase enzymes.
Epigenetic marks on DNA, such as 5mC and histone modifications, can now be added or removed using CRISPR technology. CRISPR-based eGE uses a catalytically-dead Cas9 (dCas9) fused to epigenetic enzymes or their catalytic domains. Provided with an sgRNA, the dCas9 will bind to the target, transporting the epigenetic enzymes to the site for transcriptional reprogramming.
For example, if expression of a gene has been repressed due to removal of the histone 3 lysine 27 acetylation (H3K27ac) by deacetylase enzymes, dCas9 fused to P300 can restore the H3K27 mark, leading to upregulation of the gene. eGE offers many exciting research and therapeutic possibilities, however, the ethical considerations remain largely unexplored.
Restoration of histone acetylation rescues anxiety and alcoholism
During adolescent development, there’s a lot going on in the amygdala, which is the part of the brain involved in making responses to fear and anxiety. Complex changes in gene transcription, epigenetic modulation, and the connectivity of the limbic system take place. Because the limbic system plays a critical role in behaviour and emotion, any interference in its development can have significant effects on behaviour later in life.
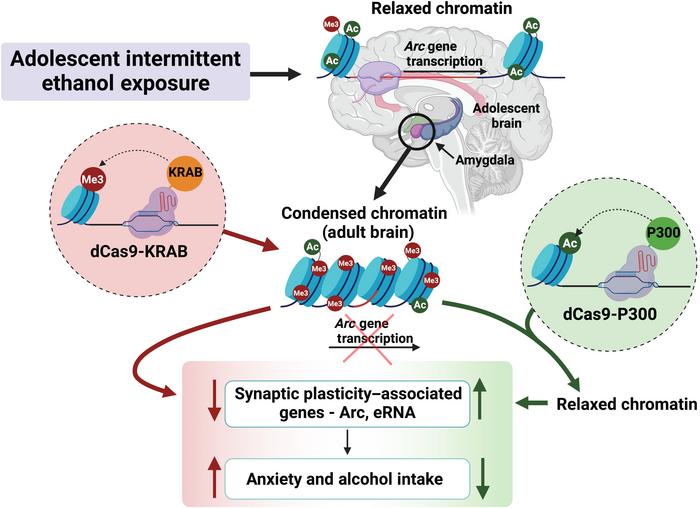
Pandey and his team hoped to use eGE to finally prove that epigenetic marks induced by adolescent binge drinking are a causal factor in AUD and anxiety. They were interested in the regulation of activity-regulated cytoskeleton-associated protein (Arc), a key regulator of synaptic plasticity and organisation expressed in the amygdala. Arc has a key role in high-order cognitive processes and addiction behaviours and operates under complex epigenetic and post-translational regulation. Its enhancer region, the synaptic activity response element (SARE), can be epigenetically repressed due to alcohol exposure during adolescence.
»The epigenome plays a very important role in brain maturation. If there is adolescent alcohol exposure, it can perturb the epigenome and there is genetic reprogramming. We found that this really does occur and it persists into adulthood - it causes anxiety and promotes excessive drinking,« Pandey explains.
Importantly for the study, SARE is conserved from rodents to humans, making it an ideal target for a translational study between rats and humans. Adolescent alcohol exposure is thought to reduce Arc expression in the amygdala of rats in a few ways, including through removal of the histone 3 lysine 27 acetylation (H3K27ac) epigenetic mark, and addition of H3K27 trimethylation (H3K27me3) at the SARE enhancer. The team hypothesised that CRISPR-dCas9 epigenetic editing could be used to reverse these effects in adult rats.
The first step was to get adolescent rats drunk, resulting in reduced Arc expression and increased anxiety (as measured by established tests) and alcohol consumption when the animals reached adulthood. To rescue Arc expression, the team employed dCas9 fused to the catalytic domain of P300, a human histone acetyltransferase that catalyses H3K27ac, with sgRNAs specific to the Arc SARE site.
The results were astonishing: the treatment reduced anxiety and drinking behaviours back to the levels seen in control rats which were never exposed to alcohol during adolescence. Just as exciting was the finding that dCas9-P300 produced no effects at any of the predicted off-target sites.
Chromatin looping, enhancer RNAs, and negative elongation factor interactions
Another interesting result was the broader effect of dCas9-P300 epigenetic editing on other genetic components. Regulators of gene expression, enhancer RNAs (eRNAs) are transcribed from either side of the Arc SARE enhancer site. Studies have demonstrated that eRNAs can bind to negative elongation factor (NELF), a protein that halts RNA polymerase II at the Arc promoter and inhibits its transcription. By binding to NELF and removing it, eRNAs can increase transcription of Arc.
Pandey and his team found that treatment with dCas9-P300 targeted to the Arc SARE site restores expression of eRNAs and blocks NELF from binding to the Arc promoter, contributing to its increased expression. Not only that, but the addition of H3K27ac at the SARE site by dCas9-P300 also causes addition of this same epigenetic mark, and reduction in the repressive H3K27me3 mark, at the promoter of the gene.
This result was suggested to be caused by enhancer-promoter chromatin looping, so the team employed a chromatin conformation capture assay to test it, analysing the interaction between the promoter and the SARE site, as well as other regions of the Arc gene. The assays confirmed that the interaction between the promoter and enhancer sites is reduced in rats that were exposed to alcohol during adolescence, and that treatment with dCas9-P300 can increase it through changes in chromatin conformation.
»It was surprising - I didn’t expect that if you increase histone acetylation 7 kb above the promoter that you will also increase histone acetylation at the promoter itself. We found that chromatin looping occurs, which is really very fascinating – you manipulate the epigenome and you see this chromatin architecture is changed as well,« Pandey remarks.
Proving that Arc SARE repression causes anxiety and alcoholism
To prove a direct causal link between the epigenetic reprogramming of SARE and adult anxiety and AUD, the team also needed to use dCas9 to mimic this change in adult control animals. Pandey theorised that using dCas9-KRAB to deposit H3K27me3 at SARE could induce these behavioural disorders even if the rats had never been exposed to alcohol during adolescence, proving that it is indeed downregulation of the Arc SARE site - and not other factors arising from adolescent alcohol exposure - that cause these disorders.
To do so, they used dCas9 fused to Kruppel-box associated domain (KRAB), a human zinc finger protein that acts as a transcriptional repressor; dCas9-KRAB increases histone methylation marks, repressing target genes.The theory proved correct – treating control animals with dCas9-KRAB targeted to the Arc SARE site decreased Arc expression and increased H3K27me3, resulting in anxiety-like behaviour and increased alcohol consumption in the rats. No off-target effects were observed, and again, the role of enhancer RNA and NELF were clear.
»We see there is a reduction in eRNAs, and because eRNA levels go down, NELF binding goes up, so we see that there is downregulation of Arc. And of course, we see anxiety and drinking. We got very excited that there is a bi-directional effect – we can turn it on and off. And through that epigenetic mechanism, we can regulate these behaviours,« Pandey elaborates.
Follow-up studies and hope for other disorders
Pandey says his team are now gathering data in several different follow-up studies, which he says must be prioritised before any potential therapeutic option can be offered. These include repeating the present study on female rats to determine sex-specific differences, defining the levels of adolescent binge drinking that lead to epigenetic changes in the amygdala, further exploration of how long the treatment will last, and the effects of epigenetic modifications on other genes expressed in the amygdala.
Professor Stanley Qi of Stanford University, a pioneer of dCas9 systems is also enthusiastic about the results of this study and the implications for the broader field of epigenome editing. Qi’s lab recently published a Nature Cell Biology paper describing a hyper-efficient dCas12a variant, which can be used for multiplexed transcriptional regulation of target genes.
»I find the work very exciting and reinvigorating. This points to the power of epigenome engineering to address a broader class of complex diseases that may not be easily tackled by simple genome editing. The demonstration of alleviating neuropsychiatric disorders including anxiety and alcohol drinking is very exciting. While the technology can be further tested in more complex disease models and can be further improved to incorporate new developments from CRISPR and epigenome engineering, it points out a bright future of applying precision epigenome editing to address diseases we hadn’t imagined decades ago,« Qi remarks.
In addition to the neuropsychiatric disorders explored in this study, Pandey is hopeful that epigenetic editing will offer therapeutic benefit for other conditions, as well as helping to unravel causes of neurological disorders and providing better models for the study of these:
»If you look at depression, trauma, other drugs of abuse, they all affect the epigenome. So this epigenetic editing approach can be applied across many diseases of the brain, and I think it has a lot of future potential.«
Link to the original paper in Science Advances:Targeted epigenomic editing ameliorates adult anxiety and excessive drinking after adolescent alcohol exposure.
Rebecca Roberts, Ph.D. is a molecular biologist and science writer/communicator based in Queensland, Australia.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







