Epigenome Editing
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Hello and welcome back to Making The Cut. Today, we publish part 20 of this series! And to celebrate that, we have a nice special episode on epigenome editing.
This is going to be a nice one, as we will be exploring a brand-new way of doing genome engineering.
In the previous episodes, we told you how scientists have been squeezing their neurons out to come up with ways to edit the genome without breaking the DNA.
As we saw, cells can be quite touchy about that sort of thing. The genome integrity surveillance system is not happy when the DNA is breaking apart and it may commit cells to an anticipated retirement or premature departure.
So long, gene-edited cells!
As we mentioned in our last episodes, this issue led to the development of new technologies like base editing, prime editing and click editing. However, despite their proven potential to do great things, those of you who tend to be quite meticulous may have noticed that these technologies do still require partially cutting the DNA.
Sure, a single-strand break is milder than cutting both strands, but still, you can’t call it a DNA-break-free technology, can you?
So, let’s all meet Bachelor number four….(drum roll, please)
Epigenome editing!
We know, it sounds like we just made that up, so let’s take a few steps back.
Epigenetics: punctuation saves lives!
In the past, we told you that DNA can be considered as a book containing all the necessary information to execute a given action, like producing a certain protein. Thus, altering the way the information is written - which we call a mutation - may disrupt the action performed.
However - like a book - the DNA is not just an endless stream of words. Unlike some people – who seem to have developed the unpleasant ability to talk without the need to draw breath - a book has punctuation: full stops, commas and even exclamation points!
And so does the DNA. Let’s give an example:
“Let’s eat, Timmy!”
“Let’s eat Timmy!”
In the first sentence, we have a happy family gathering around for supper.
In the second case, a kid just went missing in the neighborhood…

This also happens in our DNA. Let’s see what we mean exactly.
Chromatin: the genome’s punctuation
You may recall that DNA is not a loose filament in the nucleus, but rather it is organized as a bundle wrapped around a protein core known as histones, forming a complex called chromatin.
Having said that, the way histones entangle the DNA is not the same in each part of the DNA length. It can be packed more or less tightly. These two types of chromatin are called euchromatin and heterochromatin respectively.
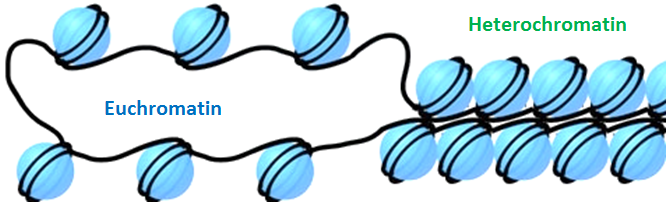
Chromatin plays a major role in gene expression, as modifying the packaging shape can bring different DNA regions closer or move them apart (ref.1). Making the packaging tighter or more relaxed may also render the DNA less or more accessible to key proteins, particularly the so-called transcription factors.
All of this ultimately affects gene expression.
Have you ever wondered how each one of the 30+ trillion cells in your body may contain exactly the same DNA and yet you have different cell types with specific functions? Muscle cells, neuron cells and skin cells are genetically identical and yet are phenotypically so different!
The trick is that while all our cells have the same exact genes, they express them at different levels. Think again about punctuation: in some cells, gene A has the equivalent of an exclamation mark, which increases its expression. In other cells, the same gene has a mark telling it to keep it down or be completely silent! This is what we call epigenetics.
Thus, gene expression results from the interaction between a gene’s sequence and the regulatory mechanisms that act upon it. Interestingly, some epigenetic modifications are inherited upon cell division!
But since such modifications are still not altering the DNA, they can also be reverted.
How cool is that?
Epigenome editing: let’s modify gene expression!
Having established that the DNA sequence is not the only layer determining whether the information contained within will be conveyed, we can now jump to the fun part: playing around with epigenetics!
How can we do that?
We will try to keep the nuances of how epigenetics works to a bare minimum, but if you are eager to know more, please reach out in the comment section or drop us a message, we are geeks and we are happy to tell you more!
Suffice to say that leveraging how epigenetics and its factors work, we may be able to turn OFF or ON the expression of a gene at will.
An example is DNA methylation, a type of punctuation mark that leads to transcriptional repression (e.g. silencing) of a neighboring gene by both hindering the recruitment of transcription factors and also by recruiting additional silencing factors. Methylation is performed by DNA methyltransferases (DNMTs) like DNMT3A, DNMT3L or other factors like KRAB.
Basically, they are like Ross from Friends, no better way to picture that passive-aggressive "shushing" move!

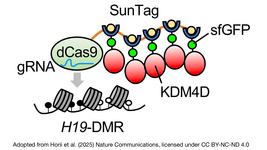
These factors or combinations of them have been fused to the catalytically dead version of CRISPR (dCas9) to create a reprogrammable protein able to direct gene silencing at a desired location in the genome!
In this case, rather than targeting an exon – as you would do with a standard CRISPR to do a KO – the dCas9-epigenetic complex is directed to a CpG island-rich region upstream of the promoter of the desired gene, which leads to heavy methylation of these region and the silencing of the gene!
However, you may also want to turn a gene on and increase its expression! We got you covered: in this case, we would attach to your dead Cas9 factors meant to boost the promoter activity, like strong transcription factors or epigenetic factors meant to relax the chromatin, instead of stiffing it! This is the case of a dead Cas9 fused to the transcriptional activators VP64, p65 and Rta…code name VPR. Yep, it sounds like VIPER, and it is great at activating the expression of whichever gene we want to target.
While still staying on Friends we can think of CRISPR-VIPER as the hyperactive Joey visiting London for the first time.

Why use epigenome editing and what we can do with it?
Ok, so now enough with the Friends references and let's see how we can practically leverage epigenome editing.
Well, we all saw how DNA damage can be a deal breaker –pun intended – when it comes to genome safety. Epigenome editing offers a real solution to that!
For example, the team of Claudio Mussolino have used it in T cells to knock down the CCR5 receptor which the HIV uses to invade our immune system! Also, the team of Angelo Lombardo has recently used epigenome editing to silence the PCSK9 gene – responsible for cholesterol accumulation and cardiomyopathies – in a mouse model, offering a new and perhaps safer alternative to Cas9-mediated PCSK9 KO for the treatment of hereditary hypercholesterolemia.
These are just a couple of examples of how Epigenome Editing may be leveraged for therapeutic application, but there is a growing number of companies – like Chroma Medicine and Tune Therapeutics – whose goal is to fully tap into the potential offered by epigenome editing to treat a different range of conditions.
So here you have it! A new way to do (epi)genome-editing without even introducing a dent to the DNA! As you see, the further the gene editing field progressed, the more safety becomes a concern. This gives a strong message, we are not any longer in the “can we do it?” phase, but rather at the step where we know can do it, and we want to do it better!
We hope you enjoyed it and we will see you in the next one!
References:
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.