Finding the Secret Ingredient for CRISPR-Cas3 Editing in Human Cells

Assistant Professor Yan Zhang of the University of Michigan recently found a cas11 gene that encodes an integral, but hidden, component of many type I Cascade-Cas3 systems.
Cascade-Cas3 gene editing has attracted attention recently for its use in engineering bacteriophage for more efficient phage therapy. However, the currently used Cascade-Cas3 are the only Class I, type I CRISPRs to be adapted for gene editing. All other commonly used systems are Class II CRISPRs, including Cas9 (Class II, type II) and Cas12 (Class II, type V).
As an early-career researcher, Zhang wanted to develop type I-based CRISPR tools for large scale deletions in the human genome; type I CRISPRs are the most abundant and diverse bacterial adaptive immune systems and are capable of degrading large sections of target DNA. She was successful; in collaboration with Ailong Ke’s lab at Cornell University, she provided the first example that CRISPR-Cas3 can achieve large-scale genome deletions in human cells. But this was a type I-E system with large genes, and Zhang wanted to develop a more compact version using type I-C.
In an article published last month in Molecular Cell, Zhang and her lab describe a compact type I-C CRISPR-Cas3 system for genome editing. The study throws a few curve balls, including the discovery of a hidden Cas protein that is essential for type I-B, I-C, and I-D systems to perform editing in human cells. The system is able to achieve astonishingly high editing efficiencies in human stem cells and has exciting potential applications that may also include in vivo delivery thanks to its compact size.
»We can potentially use this system to create large-scale deletions to model certain genetic diseases, to delete DNA viruses from the human genome, or for selective killing of cancer cells – our tool can be a very good killing machine,« Zhang asserts.
“We can potentially use this system to create large-scale deletions to model certain genetic diseases, to delete DNA viruses from the human genome, or for selective killing of cancer cells – our tool can be a very good killing machine”Yan Zhang
Finding a candidate CRISPR system
While Zhang was previously successful in developing a type I-E CRISPR system, she knew there was room for improvement, and she set her sights on type I-C.
»The prior versions had some limitations. Type I-E is the most complicated [type I system], with the largest number of genes and gene size. We always wondered if we could find a more compact system. Type I-C is the most compact type I CRISPR system, with fewer, and smaller, genes. Type I-E are also not the easiest to purify. Additionally, type I-E all use the same PAM, but other type I subtypes, like I-B, I-C, and I-D, have different PAMs, so they have a broader range of targets,« Zhang elaborates.
The type I-C Cascade-Cas3 system Zhang decided to investigate was isolated from the bacteria Neisseria lactamica (NlaCascade-Cas3). As a postdoctoral researcher in Erik Sontheimer’s lab at the University of Massachusetts, Zhang had examined different Neisseria genomes and found what she thought might be a type I CRISPR system. While she wasn’t able to do any functional testing at the time, she says she maintained her curiosity until she established her own lab, as she explains:
»That was always on my mind. When my bioinformatics Master’s student Ryan Krueger joined the lab, I wanted him to use his computational power to dig [into this] - to see if all the Cas genes are present, if it’s widespread in Neisseria, and if he could predict a PAM based on naturally existing target sequences in the database. If all those factors checked out, then we could do functional tests.«
Once Ryan had established this information on the NlaCascade-Cas3 system, he proceeded with a plasmid interference assay, using plasmids to express the Cas genes alongside crRNA and a target sequence, as Zhang explains:
»In the plasmid interference assay, we use antibiotic selection to demand co-presence of all these plasmids together - if there is a deficiency, then that means there is active CRISPR functionality.«
NlaCascade-Cas3 achieves robust editing in human embryonic stem cells
The study was full of surprises. The first was the incredibly high editing efficiencies achieved when the NlaCascade-Cas3 purified from bacteria was delivered to human embryonic stem cells as a ribonucleoprotein (RNP) complex (Figure 1). In some cases, editing efficiencies were as high as 95%.
»I was at a conference when my lab members emailed me the results of editing in stem cells with this system. Stem cells are always a little more difficult to get super high editing efficiencies. In our previous paper we used the same cells, so I was used to seeing those relatively low numbers – the best was about 13%. So when my student told me they got 50%, that was really great,« Zhang recalls.
The team wasn’t going to stop there – once they realised how high the editing efficiency was with the NlaCascade-Cas3 system, they wanted to explore multiplexed editing.
»Sometimes you really need co-disruption of two or more alleles in the same nucleus so that motivated us to try multiplexing. If single-locus editing efficiency is already low, multiplexed editing is not going to happen, but because we had higher numbers [with NlaCascade-Cas3], multiplex editing was within reach,« Zhang explains.
The Cas3 nuclease has rather unusual activity – instead of creating a double-stranded break in DNA, like Cas9, Cas3 ‘shreds’ it. Cas3 is recruited and activated by the target recognition complex known as Cascade (CRISPR-associated complex for antiviral defense), which in Neisserisa lactamica is thought to be made up of the Cas5, Cas8, and Cas7 subunits. Cascade recognises the target sequence and forms the R-loop for editing, after which Cas3 is recruited and begins degrading the DNA in one direction, deleting large chunks of DNA.
»The deletions are commonly within the 30kb range. We still don’t know what causes Cas3 to stop shredding. We haven’t pinned down the trigger yet - it’s a seemingly random process to us. One of the most important focuses now is understanding that and finding ways to control it. We may also be able to find naturally existing orthologues that offer different processivity,« Zhang remarks.
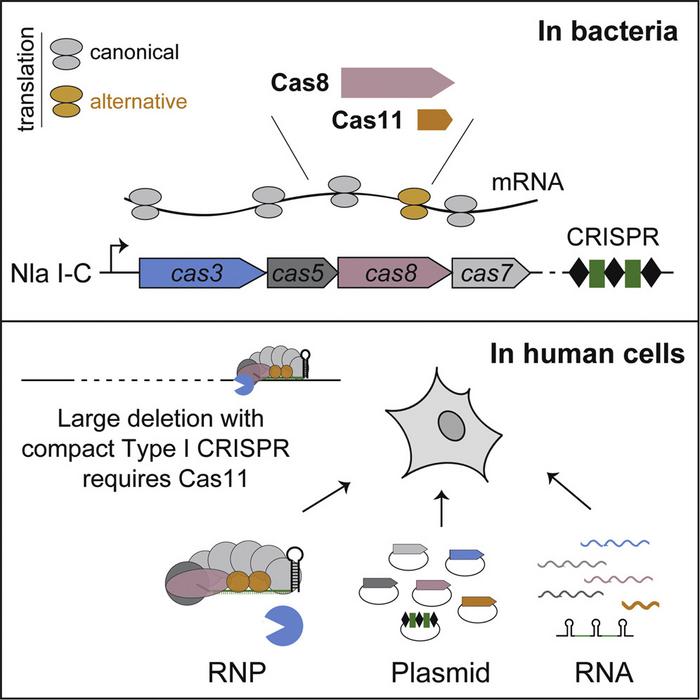
A major hurdle leads to an unexpected discovery
After achieving such robust editing in human embryonic stem cells with the purified proteins, Zhang and the team started thinking about writing up the paper. Before doing so, Zhang insisted that they develop a plasmid-based platform for the NlaCascade-Cas3 system.
»We can’t demand all users to go through the process of purifying proteins and reprogramming the guides – DNA-based delivery would be ideal for some users. But then we started running into problems with unexpectedly low editing efficiencies,« Zhang notes, who then continues:
»We did months of troubleshooting. We did western blots to look at protein levels and we titrated every plasmid’s concentration, but we couldn’t figure out how to get the efficiencies above 2%. We thought that maybe the complex simply failed to assemble in human cells and that this led to weak editing efficiencies.«
Zhang was beginning to think they should cut their losses and move on until her graduate student and first author of the paper, Renke Tan, eventually solved the NlaCascade-Cas3 plasmid problem. After closely examining the Cascade protein expression bands, she noticed an extra band. Tan was determined to find out what it was, and as Zhang puts it, her curiosity paid off.
»I give credit to my graduate student - she was able to think outside the box. She looked at the protein expression gel and said – this is not clean, there’s an extra band. What is that? Are we not purifying the protein correctly? Eventually she cut out that band and sent it for sequencing and we realised it was an internal translation product. Normally, we wouldn’t even have followed up on that extra band. If it wasn’t for her, we wouldn’t have discovered this hidden gene,« Zhang explains.
That internal translation product was Cas11, and it proved to be essential for Cascade-Cas3 editing more broadly. In previously used type I-E CRISPRs, cas11 is an obvious gene, producing a separately encoded protein that forms part of the Cascade complex. In type I-B, I-C, and I-D systems, however, cas11 is a ‘hidden’ gene, requiring alternative translation from within the cas8 gene.
The problem with these systems in human cells is simply that human ribosomes are unable to recognise the internal prokaryotic translation initiation site within cas8. Zhang emphasises that a fruitful collaboration with Ailong Ke’s lab was essential for the success of the project and the discovery of Cas11 as an essential component of the system.
»I really appreciated that [Ailong Ke’s lab] brought an RNP angle to the project. Plasmids are easier to manufacture, but if we had used that to do our initial screen for I-C orthologs, we would have taken much longer to discover the usefulness of type I-C systems,« Zhang says.
Around that time, there was confirmation from an independent study published in Molecular Cell, describing the same internal Cas11 production phenomenon in type I systems in other bacteria. Zhang’s team were relieved to see that this seemingly strange occurrence was more widespread in the bacterial world. They finally had the answers to the type I CRISPR problem.
» It was very satisfying. After we added a plasmid with cas11 we were able to get back to editing efficiencies of 20-30%. Then we tried a few other compact type I-B, I-C or I-D systems - they showed varying degrees of activity, but they are functional in a cas11-dependent fashion. Everything from there went quite smoothly,« Zhang says.
The next challenges: off-target editing and real-life applications
Zhang says a major challenge for the type I CRISPR field is to explore and define off-target editing events so that these systems can be safely adopted for editing in human cells. So far, it seems likely that off-target activity is fairly minimal. A 2019 Nature Communications paper examined a type I-E Cascade-Cas3 system from E. coli, performing whole-genome sequencing of edited cells and finding no obvious off-target editing. Zhang and her team hope to explore this in more detail:
»We want to do a more targeted approach. We want to predict the top 20 to 50 ranking sequences with some similarity to the target sequence, then go in and design PCR experiments to check for large deletions and potential chromosomal rearrangements. We also want to perform deep sequencing to check for potential indels. Right now, we’re doing some mismatch tests to comprehensively understand the type I CRISPR mismatch tolerance rules in human cells so that we can predict possible off-target sites, and then really profile those loci,« Zhang says.
Zhang and her team are excited about applying their robust NlaCascade-Cas3 system to a variety of research questions, including targeting solid tumours. Currently, they’re collaborating with other labs to use the system to study non-coding elements that regulate embryo development and stem cell differentiation, and to create large deletions to model certain genetic diseases.
»The highlight of the research for me was making unexpected discoveries. All earlier works, by us and others, used type 1E CRISPR systems. So I thought, there must be a reason why this is the only subtype that works. In retrospect, that’s because I-E systems have cas11 as a well-annotated, obvious gene, not a hidden one. It answered a really long-standing question in my mind,« Zhang concludes.
Link to original article in Molecular Cell:
Cas11 enables genome engineering in human cells with compact CRISPR-Cas3 systems.
Rebecca Roberts, Ph.D. is a molecular biologist and science writer/communicator based in Queensland, Australia.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







