How CRISPR Cuts the DNA and What the Cell Does About It
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
In the previous episode, we left you with the main idea that the first iteration of CRISPR is no more than a molecular scissor, and how the “editing” or the “engineering” part of a CRISPR experiment is a consequence of the repair mechanisms engaged by the cell to make up for the cut made by CRISPR.
DNA repair mechanisms are quite complex, so in this short article, we will stick to the main gist which helps us understand the foundation of a CRISPR experiment.
We already mentioned that CRISPR cuts both strands of the DNA, creating a so-called DNA Double-Strand Break (DSB).
Now, let’s see what happens in a cell when a DSB occurs.
In short, all the emergency signs light up and a well-orchestrated cascade of events starts.
It is a controlled panic.
This controlled panic takes the name of DDR or DNA Damage Response.
But first things first: DSB detection!
A group of kinases (ATM/ ATR)– proteins adding a phosphate group to other proteins –can detect a DSB, which will lead them to phosphorylate H2AX. This is a peculiar type of protein around which the DNA is wrapped around – a.k.a. histone – which acts as a beacon to signal that a DSB has occurred.
Yes, imagine it like a bat signal.
What happens afterward is a series of events that lead to two main outcomes: temporary cell-cycle arrest and DNA repair.
The block of the cell cycle – at defined points also called “checkpoints” –is instrumental to a) facilitate the repair of the DNA damage and b) prevent the DNA alteration from being spread to the daughter cells.
So, the DSB has been detected, the bat signal has been turned on and the cells are on stand-by waiting for repair.
What happens next depends on several factors involved in the repair of the DSB, which we will mention in a more advanced stage of this series.
1. DSB-ends are protected: a complex of factors Ku70 andKu80 surrounds the ends of the DSB and protects them from further “injuries”. What can happen at this point is
i) the two ends as they are can be immediately stitched back together d by the Ligase IV (Lig IV)
ii) the two ends are not compatible. This can be due to a variety of alterations following the DSB. For this reason, the two ends need to be resected – by a complex where Artemis is one of the main factors – in order to render the two ends compatible and “ligable” together. During this step, mutations like insertion or deletions of DNA bases – aka indels – may happen which may have consequences. This mechanism by which the two ends are simply joined together is also called Non-Homologous End Joining (NHEJ).
2. DSB-ends are processed and the homologous chromosome is used as a template: contrary to the first scenario, in this case the ends of the DSB are resected in only one of the two strands in both directions by a complex of factors called MRN and CtIP. This leaves two single-stranded DNA ends. These single-stranded ends are then coated by a group of factors – mainly RAD51 which “guide” the single-stranded ends towards its matching sequence on homologous-chromosome. The homologous chromosome is then used as a blue-print to faithfully recreate the information lost as a consequence of the DSB. This DNA repair mechanism is called Homology-Directed-Repair (HDR), and with it, no mistakes are introduced during DNA repair. However, this mechanism is mostly active in dividing cells, and thus cannot be the only mechanism.
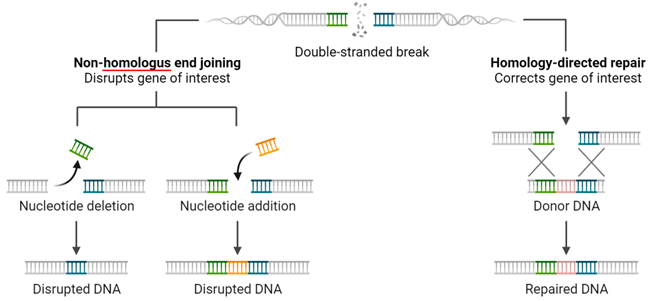
Ok, let’s take a deep breath. We left the hard part behind.
Now, let’s talk some fun. How do we use what we read so far to do genome editing?
Let’s focus for the moment on NHEJ only. We will talk about HDR applications later.
If a DSB is repaired via NHEJ it may happen that mistakes – or indels – are introduced.
Indels occurring within the coding region of a gene – aka exons – may cause an alteration of the way the information contained within the gene is interpreted. Meaning that the “open reading frame” could be altered.

Particularly, indels may shift the reading frame leading to changes in the way it reads. This can result, for example, in a premature stop codon, which may lead to a very short and unstable mRNA or to a truncated and unstable or non-functional protein. This may eventually result in the loss of function of the gene.
By connecting the dots, we get to the point where if we make CRISPR cut within the exon of the desired gene, by alternating its reading frame as a consequence of the indels introduced during the NHEJ, we may abrogate the gene's function. Or – using the standard jargon – we may Knock-Out (KO) the gene.
Now, how do we do this, practically? How can we direct CRISPR toward the desired gene?
Don’t worry, we are not going past anything. Rather give yourself some time to let this sink in and stay tuned for the next episode!
As always feel free to comment, make suggestions, and reach out to us!
Link to the previous episode here.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







