How Does CRISPR Work?
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Step by step we are getting a clearer picture about CRISPR/Cas as a genome editing tool.
So far we learned that a) the CRISPR/Cas system's main function is to cut the DNA and create a particularly severe lesion called DNA-double stranded break (DSB), b) cells have in place a complex DNA Damage Response (DDR) system that springs into action immediately as soon as it detects a lesion, and c), by leveraging the DDR it is possible to achieve desired genetic alterations.
The next questions we will answer in the coming articles are how we target CRISPR/Cas toward a desired gene and where exactly - within the gene of interest (GOI) - should we direct it?
Buckle up, we are finally getting to the "engineering" part of the matter.
Finding the target
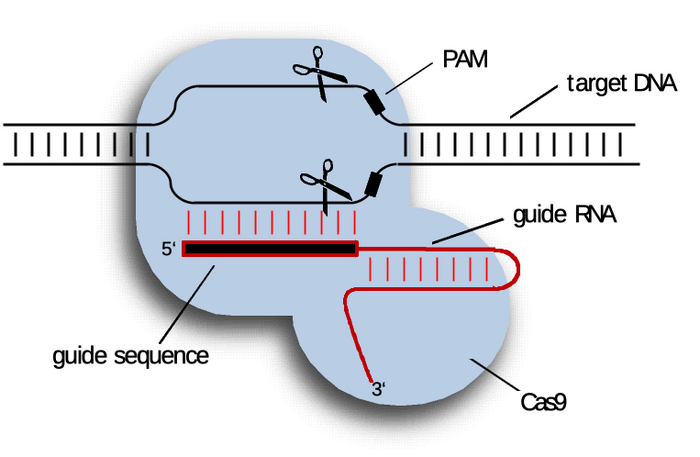
Till now we defined CRISPR/Cas as a "molecular scissor". This scissor belongs to the Cas protein family and the most known is Cas9. However, this scissor is not the only part of the CRISPR/Cas9 system. Because being able to cut is important, but knowing where to cut is the crucial part. For this, the Cas9 protein is equipped with a search engine that defines which section of the DNA will be cut.
Imagine that search engine like the one you use in Word, the classic CTRL+F. The search engine takes the input that you write and looks all over your text to find whether the text you are searching is in there. Well, Cas9 has the ability to do the same and can look for any input you give it in all of the cell’s DNA. But how do we give this input? For that we have to use an RNA molecule called a single guide RNA (sgRNA).
So every time you hear about CRISPR you have to imagine a binary system made of the protein scanning the DNA - Cas9 - and the input guiding it, the sgRNA.
We will skip past the whole CRISPR/Cas biology (for now) and we will narrow it down to the fact that the sgRNA has two main units 1) a scaffold that binds it to the Cas protein and 2) the spacer sequence, which defines a complementary region on the genome which will be targeted by the Cas.
So you can already appreciate the beauty and ease of use of the CRISPR/Cas system: by changing the sequence contained within the spacer (the input for the search engine), you can redirect the Cas protein towards a desired target.
CRISPR is cheaper and easier to design than other gene editing tools
If this does not seem like a big deal to you, imagine that before CRISPR/Cas, the only way to target a protein to cut the DNA was to use ZFNs and TALENs, proteins that recognized a target sequence through protein-DNA interactions. This meant that every time you wanted to target a different sequence, you would have to engineer the binding domain of a ZFN or TALEN, essentially making a new protein to target your sequence. This process was complex, lengthy (weeks to months) and expensive. It would be like having to build from scratch a search engine that only detected the word “Saturday”.
With CRISPR/Cas, the only thing needed to target a new sequence is to change the sequence of the sgRNA, which is a matter of days or even one click if you get it synthesized by one of the many providers out there!
The PAM sequence is essential for CRISPR to scan the DNA
All right then, can we actually target any place we fancy in the genome as the advertisement says?
If you are used to commercials, you know that “limitations” may apply.
Stay calm, it is not that bad.
The limitation in this instance is called the Protospacer Adjacent Motif, or PAM for friends. A PAM is a defined sequence that constitutes the first point of interaction of the CRISPR/Cas complex with the DNA.
You can literally picture the CRISPR/Cas complex scanning the DNA for its PAM and then trying to match the sgRNA spacer with the sequence right next to this PAM. If the two sequences match (are complementary to each other) the Cas protein progressively unwinds the DNA and the DNA sequence – thanks to its interaction with the sgRNA spacer sequence – is accommodated within the catalytic domains of the Cas protein which triggers the DSB. Basically, the recognized target sequence is placed between the two blades of the Cas scissor, and… Snip!
Interestingly, Cas9 proteins from various bacteria species recognize different PAMs. In the case of the most used Streptococcus pyogenes Cas9 (SpCas9) the PAM is a trinucleotide motif NGG (and with a lower efficiency also NAG), where “N” can be any nucleotide. Therefore, any sequence in the genome can be a target for SpCas9 provided that an NGG is right next to it!
While PAM availability can be seen as a limitation for the applicability of CRISPR/Cas9, researchers worldwide have worked hard to mitigate it by researching Cas9 – or even other Cas”n” – proteins derived from other bacterial species with different PAM specificities, and/or engineering the current SpCas9 to have a more relaxed or virtually no PAM requirement. But this is a story for a more advanced stage of this series of articles!
Take home messages:
- CRISPR/Cas is a binary system made of: the DNA nuclease, Cas protein and a guide-RNA whose spacer sequence determines the target
- CRISPR/Cas can be directed virtually towards any desired target within the genome provided a PAM motif is there, by providing a sgRNA with a spacer complementary to the target sequence.
All right, that's it for today folks! In the next chapters, we will see how to design the guiding unit of CRISPR/Cas and how the intended application affects where the CRISPR/Cas is targeted.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







