New Hope for Cancer Immunotherapies: CRISPR Knockout of a Chromatin Remodelling Factor Reverses T Cell Exhaustion

T cell exhaustion is the bane of any cellular immunotherapy – terminally exhausted T cells fail to persist and proliferate in vivo and are unable to clear tumours. Despite being well characterised, a solution to the problem of T cell dysfunction has remained elusive.
New evidence published in the journal Cancer Cell has identified chromatin remodelling factor Arid1A as a key regulator of T cell dysfunction. The result of a collaboration between Stanford University, University of California San Francisco, the Innovative Genomics Institute, and the Memorial Sloan Kettering Cancer Center, the study demonstrates that CRISPR-based knockout of Arid1A reverses the exhausted state of T cells in vitro and in vivo.
Principal investigator Ansuman Satpathy, a physician scientist and assistant professor at Stanford, says the results of this study - along with recent work from other labs – offers long-awaited hope for the broader field of T cell immunotherapy:
»For a long time now, we’ve been describing the phenomenon of T cell dysfunction and the molecular aspects of T cell dysfunction. But now, we’re finally able to hit the target of reversing and preventing T cell exhaustion. I believe that this is an exciting advance in our understanding of this fundamental process.«
Terminal exhaustion limits the success of adoptive T cell therapies
Satpathy has spent most of his career elucidating the genetic wiring of immune cells and their responses to pathogens and tumours. Forming his own lab not long before the beginning of the global COVID-19 pandemic, Satpathy’s recent work has focused on understanding T cell exhaustion.
Exhausted T cells display poor responses to tumour antigens, and limited proliferation and persistence in vivo. With high expression of inhibitory receptors and low levels of effector proteins, they are typically associated with poor clinical outcomes.
Previously thought to be caused simply by the upregulation of inhibitory receptors on otherwise healthy cells, a slew of evidence published in the last five years has revealed that exhausted T cells are in fact a completely distinct lineage of T cell, bearing complex transcriptional and epigenetic changes resulting from chronic antigen stimulation of the T cell receptors (TCR) expressed on their surface. Satpathy emphasises that exhaustion is as distinct a state as any other T cell subtype, such as regulatory T cells.
This cell state is now well characterised, however, the genetic and molecular mechanisms underpinning the process of T cell exhaustion remain unknown. Satpathy and his team aimed to address this key knowledge gap using high-throughput genomic technologies, with the hope of improving cellular immunotherapy outcomes for patients.
»T cell exhaustion is one of those processes that seems to be limiting in many forms of immunotherapy. That's why we tried to solve this problem - what is the molecular state of T cell exhaustion and what are the genetic drivers of this process? And can we actually manipulate that, not only to prevent exhaustion, but to make the T cells actually function better and longer?« Satpathy says.
Genome-wide CRISPR-Cas9 screens converge on chromatin remodeling factors
To identify the molecular and genetic determinants of T cell exhaustion in vitro, Satpathy and his colleagues, including first author Julia Belk, developed a scalable chronic TCR stimulation assay that would recapitulate the exhausted state in T cells. They then subjected exhausted T cells (TEX) to genome-wide CRISPR-Cas9 screens, with some surprising results.
»Even from that first in vitro genome-wide screen, what was striking to us was that epigenetic factors were highly enriched in the hits. That was our first clue that epigenetic factors were key, more so than others we might have predicted. I think this really shows the power of unbiased genomic screening - we might have predicted some transcription factors or particular epigenetic factors, but what we actually saw was this huge enrichment of epigenetic factors that we hadn't predicted. The next question was, can we improve T cell persistence by deleting any of them?« Satpathy explains.
What followed was a series of iterative, smaller-scale in vitro screens to narrow down the search for a potential target to reverse the exhausted state, followed by even more in vivo screens in tumour models in mice. The team used Perturb-seq, a single-cell RNA sequencing platform which allows for the analysis of large-scale CRISPR screens, to explore transcriptional profiles in the cells following genetic perturbation.
The iterative screens continued to home in on epigenetic regulators, particularly the inositol requiring mutant 80 (INO80) complex and the evolutionarily conserved switch/sucrose non-fermentable (SWI/SNF; cBAF) family of mammalian chromatin remodelling genes.
To cover all bases, the team explored both acute and chronic T cell stimulation, investigating which factors could improve the T cell state across both categories. As Satpathy elaborates, the criteria to select their final target were extensive:
»The T cells would have to perform well in an in vivo tumour response, and there are many hurdles that a T cell has to pass in order to do that. It has to engraft, survive, find the tumour, enter the tumour microenvironment, and deal with all the suppressive factors that are in the tumour microenvironment. After all that, it needs to recognise the tumour antigen, and then proliferate and survive there.«
Arid1a: a key regulator of terminal exhaustion in T cells
After an intensive search, the team found such a target: AT-rich interaction domain 1A, otherwise known as Arid1A. This geneis involved in transcriptional regulation through chromatin remodelling in both enhancer and promoter regions of genes. Perhaps not surprisingly, mutations in SWI/SNF genes including Arid1A are found in around 20% of characterised tumour genomes, making them the most frequently mutated chromatin remodelling factors in human tumours.
CRISPR-Cas9 knock out of Arid1A resulted in marked improvements in T cell performance in vitro and in vivo – persistence was enhanced, expression of inhibitory receptors such as PD-1 was reduced, and anti-tumour responses were increased. Notably, survival of mice that received Arid1A-depleted T cells was significantly increased compared to mice that received control cells. The team were understandably thrilled with these novel findings.
»We were excited that we were understanding something new about the process of T cell exhaustion, and that there are many genetic regulators to this process that people hadn't suggested before. In terms of translational T cell engineering, we were also really excited that the deletion of one of these factors could make the T cell perform better in all of these different situations,« Satpathy comments.
The study also examined CRISPR depletion of Arid1A in primary human T cells, followed by testing in vivo in a xenograft mouse model of melanoma. The improved persistence of the edited human cells in the tumour-bearing mice demonstrated that Arid1A is indeed an ideal target for clinical translation.
»Validating this in the human T cells was really the ultimate test for us - proving that we can translate this from mouse to human, and that it’s conserved in human T cells. That was especially exciting,« Satpathy notes.
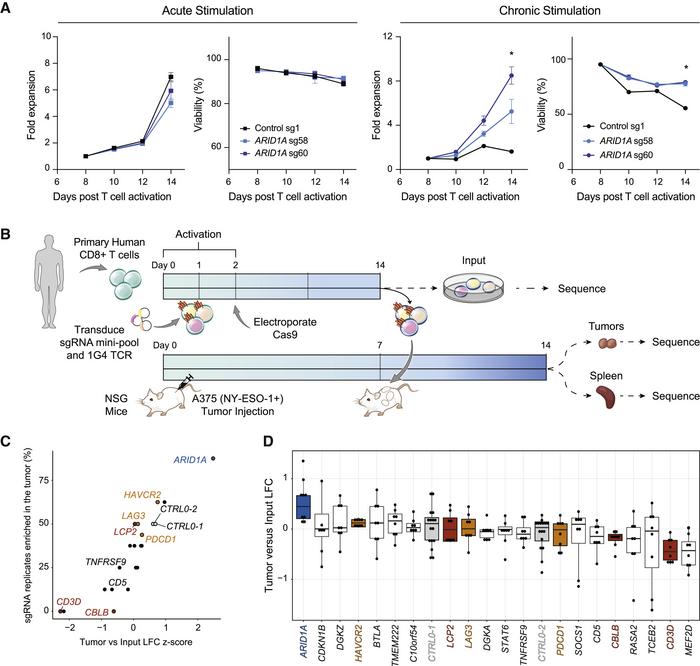
Finally, the team explored the epigenetic landscape of exhausted T cells following Arid1A perturbation to further elucidate the function of this gene. Analysis of chromatin accessibility at transcription factor (TF)-binding sites showed that not only does Arid1A depletion make exhaustion-associated TF motifs significantly less accessible, but it also makes TF motifs associated with effector T cell function more accessible.
Can CRISPR editing further enhance T cell therapies for cancer?
CRISPR editing is already being used to generate improved cellular immunotherapies - for example, to knock-in chimeric antigen receptors (CARs), to knock out the endogenous TCR, and to perturb genes that contribute to host immune rejection. The results of Satpathy’s study suggest CRISPR-based manipulation of Arid1A could soon become an exciting avenue for engineering T cell therapies, either through complete gene knockout or epigenome editing.
Epigenetically speaking, exhausted T cells taken from the tumour microenvironment bear striking similarity to those resulting from chronic viral infection, making this cancer-focused study applicable to other disease contexts.
Assistant Professor Glenna Foight, of Baylor College of Medicine, was not involved in the study but believes these findings are significant for the field. As a specialist in protein engineering and synthetic biology at the Center for Cell and Gene Therapy, Foight’s own laboratory develops molecular interventions to improve the performance of immune cell therapies.
»This study illustrates the power of chromatin remodeling factors to exert global effects on a complex, epigenetic cell state like T-cell exhaustion. Knockout of cBAF subunits had a pleotropic effect that combined upregulation of multiple beneficial effector molecules, receptors, and transcription factors with downregulation of genes that promote exhaustion. Many of these individual gene targets have been modulated in recent years to successfully improve CAR-T cell resistance to exhaustion, so the power of a single target like Arid1a that combines their effect is potentially very significant for the increased efficacy of cellular immunotherapy,« Foight comments.
As is the case for many genome-wide screening experiments, this study has spawned a variety of intriguing follow-up investigations that the team and their collaborators are currently working on, as Satpathy concludes:
»Arid1A was the best hit, but many of the other top hits were unknown genes. We only really scratched the surface in this paper, of all the other hits... I think some of them may be just as promising as Arid1A.«
Link to the original article in Cancer Cell:
Rebecca Roberts is a molecular biologist and science writer/communicator based in Queensland, Australia.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University