Off-Target Effects and Where to Find Them
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Last week, we discussed how CRISPR is being used in the real world for treating patients, and looked at some examples of CRISPR therapies that are close to being approved, as well as the first approved CRISPR therapy ever!
That was all good and fun, but we know that what you really want is to learn how to do CRISPR yourself. And today we’re going back to it with a deep dive into one of the most complex topics in the CRISPR field: Off-targets!
Off-targets and their dangers
But first of all, we should explain what an off-target is. If you remember, in Part 3 of this series we described the CRISPR/Cas9 system as a sort of “search engine” that can find any word we want in the long text that is our DNA. Then, the Cas9 protein can cut the DNA at the location of that “word”, which is going to be our target. However, this search engine is not as precise as we would like to think. Indeed, it can accidentally find words that are very similar to our target but differ from it by a letter or two. For example, if we’re looking for the word “Though”, our CRISPR/Cas9 system can get confused when it finds words like “Through”, “Thought” or “Tough”.
You can relate, can’t you?
But what happens if a wrong “word” is recognized? Well, that’s when trouble starts. Because, since CRISPR/Cas9 thinks it found the right word, it will cut the DNA at that location. This is what we call an off-target. We can tell you right away, most of the time cutting the DNA at the wrong place is not a good thing. For many reasons.
The first reason is that if CRISPR/Cas9 cuts the DNA in the coding sequence of a different gene, or its regulatory regions, we could alter or disrupt the expression of an unintended target, which may create a confounding element in our experiment and potentially bias our observations.
However, worse things can happen. One of these you can already imagine: what would happen if CRISPR/Cas were to accidentally target an essential gene? Like one of the genes that monitor the “behaviour” of our cells. If such a gene is accidentally knocked out, the edited cells could growing and dividing faster than the others. Like a cancer cell would do. In that case w would obviously have quite a problem in our hands. But wait, things may get even more complicated and dangerous!
For example, imagine that CRISPR has cut the DNA at two different locations (our target sequence and one off-target sequence). This would be like having an instruction manual ripped in three parts. If we wanted to put that manual back together quickly, the probability of us doing that wrong would be pretty high. And what would happen if we mixed up the parts? We could end up with a text that made no sense, right?
Well, that can happen in our DNA as well and can cause the formation of new genes with cancerogenic potential. And that’s not everything. In some cases, maybe we put together the two ends of the manual and then realize we forgot the middle part. That would lead to the deletion of a large section of DNA, and the genes included in it. Not great.
Luckily, our cells are pretty good at detecting when something is wrong with their DNA. Especially if there’s a lot of damage in the DNA (and several cuts do count as a lot of damage). We are sure you remember from the second article our great ally the DNA Damage Response (DDR). It is always on our side, even if from time to time we trick it to CRISPR the cells. This DDR includes several security systems that stop it from reproducing and try to fix the damage.
But what if the damage is not easily fixable?, we hear you ask. In that case, the cell will decide to self-destruct, in the cellular death process known as apoptosis. This makes sense in the context of a larger organism, as we can just get rid of damaged cells and replace them. But it does not bode well for our experiment when our edited cells kill themselves, does it?
You can hear the off-targets laughing evilly at us. Let’s see how we can catch them before they can do any harm!
Studying off-targets
Okay, so off-targets=bad, we got that. But what do we do to avoid them? Is it even possible to do so? As we explained in Part 4, where we learned how to design gRNAs, the software that we use can predict the off-targets for our gRNAs and the probability that our CRISPR system will recognize and cut them. Thus, selecting gRNAs with fewer and less common off-targets is a good way to reduce their probability.
Additionally, we can extract DNA from our CRISPR-treated cells and sequence the regions where these off-targets could occur, to quantify how much they happen. For that, we will use NGS-based methods as we explained in Part 5. This is because, thankfully, off-targets are usually not very frequent and therefore we need highly sensitive methods to detect them. NGS-analysis algorithms like CRISPREsso allow us exactly to do this.
However, not all off-targets are predictable. In addition to that, these in silico software cannot account for all the potential different experimental conditions. Among them, for example, which cell type is being used. In fact, different cell types may differ in their DDR and therefore in their ability to “self-heal” a cut in the DNA and potential off-target.
The ability to detect these low-frequency off-targets is essential both when working in a Petri dish or directly on a living organism. In both cases, we talk about millions of cells. If we are working with 10 million cells, even an off-target that happens only in 0.001% of the cases will give us 10.000 potential wrongly edited cells that may just flip and start doing some nasty things. Thus, we are going to need methods with a higher sensitivity to detect all potential off-targets.
Luckily, scientists have developed several systems that can help us with that. The lab of Keith Joung at Massachusetts General Hospital has pioneered several of these systems. The two most relevant ones are GUIDE-seq and CIRCLE-seq.
GUIDE-seq: This strategy uses a small DNA tag molecule that integrates itself very efficiently into any CRISPR-generated double-strand break (DSB). When cells are treated with CRISPR and a specific sgRNA, their DNA can be extracted and the tag can be retrieved by next-generation sequencing. Thus, any DNA region where the tag has integrated will be recognized as a CRISPR cut, either on-target – when the tag is detected at the intended region – or off-target – when the tag is detected in other genomic locations -. What’s more, the more commonly the donor molecule is found in a specific region, the more likely that off-target will be.
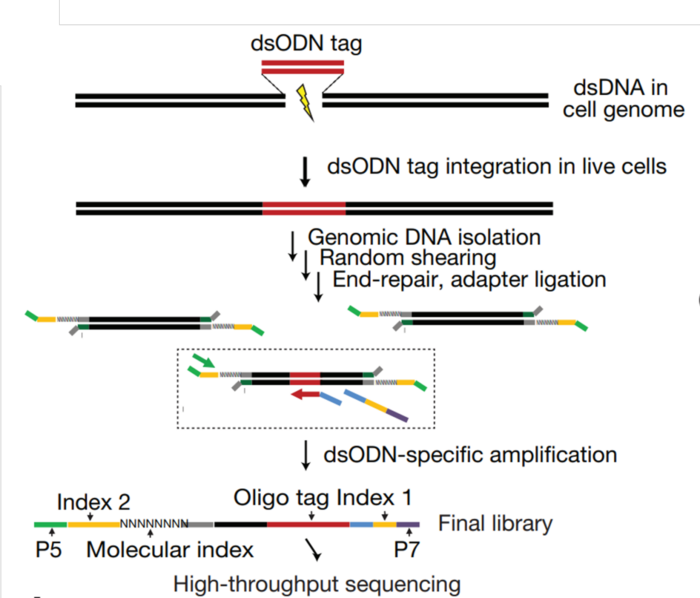
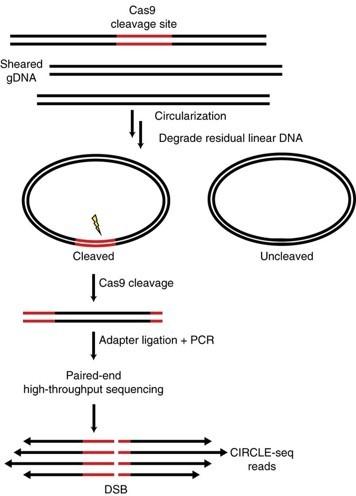
CIRCLE-seq takes that idea a step further by completely removing the need for cells. Indeed, we can measure CRISPR off-targets without even putting it into our cells! In fact, we just need a reaction tube! What CRICLE-seq does is take the cell’s DNA and break it into little fragments which are then circularized, making them into tiny circular DNA pieces. These pieces will contain the regions harbouring the on-target site and the potential off-targets. The DNA circles are then put into a tube together with CRISPR and the sgRNA. In this situation, any time CRISPR cuts, it will turn a circular DNA piece into linear DNA. The trick of this approach is that circular DNA cannot be sequenced. Instead, linearized DNA cut by CRISPR can. Thus, we will only read those sequences that have been cut by CRISPR.
This method significantly increases the sensitivity of off-target detection. In addition, the identified off-target regions can then be sequenced in depth to better quantify their editing frequency. However, it is important to note that CIRCLE-seq will present a “worst-case-scenario” picture. Since the DNA extracted from a cell is more accessible to CRISPR/Cas compared to when it is tidily packed inside the nucleus, some sites will get cut in CIRCLE-seq that may never be cut in a cell. Thus, while CIRCLE-seq is a great tool to discover potential off-targets, they need to be further confirmed with additional techniques like GUIDE-seq or the one we are about to introduce you.
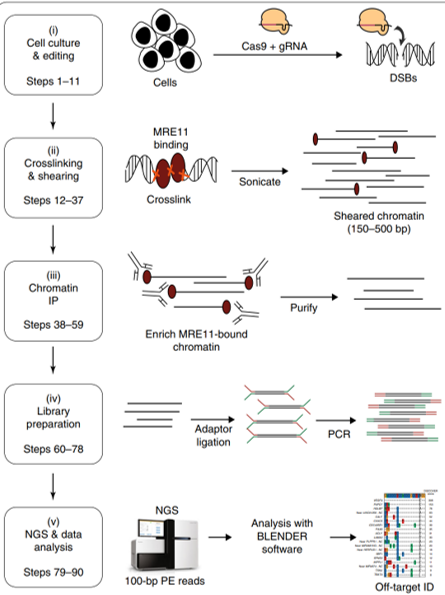
DISCOVER-seq: This method, developed by the lab of Jacob Corn, uses chromatin immunoprecipitation to isolate those DNA sites with a DSB, by immunoprecipitating the proteins that bind to those DSB. Then, these DNA sites are sequenced and identified.
Detecting unintended translocations
Wait a second, what happened to the dreadful “shredded manual” example? Can we detect such events? We were just checking your attention level. You passed.
Yes, there are methods out there that have been tailored exactly to detect the possibility that pieces of DNA belonging to different genomic locations may end-up together as a desperate attempt to repair a DSB occurring at the on-target and another one occurring at the off-target-
The most recent one is CAST-seq, developed by the group of Toni Cathomen.
CAST-seq cleverly uses a PCR-reaction based on a prey-bait strategy. It looks like we are going hunting! In this case, the bait is going to be our on-target site, and the prey is the off-target. The DNA is extracted from CRISPR-edited cells and fragmented into smaller pieces – like CIRCLE-seq for example – but it is not circularized. Instead, short DNA sequences are attached via a chemical reaction at the two ends of the DNA fragments.
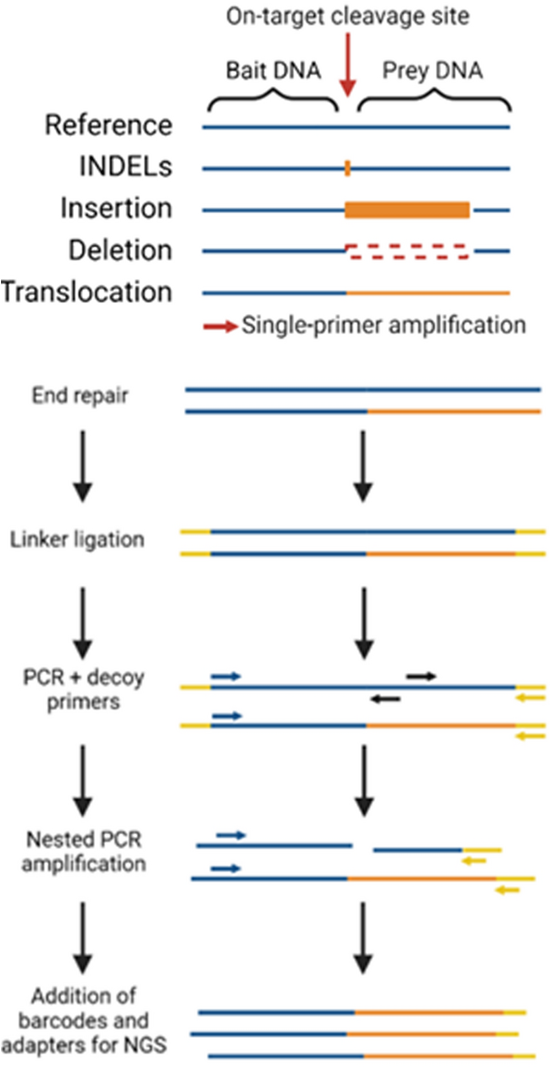
Now comes the cool part. We can design a PCR reaction where one primer binds the on-target, for which we know the sequence, and the second primer binds to the short DNA sequence that we added to the opposite side of the fragment. Now, if we run this PCR, we can amplify and sequence whatever pieces of DNA are between the two primers. If an unexpected DNA sequence is found downstream of our bait sequence, this could potentially be a piece of DNA that fused to our on-target sequence as a result of a genomic translocation.
Get the grill ready, we got ourselves a big beefy off-target!
Okay, great. Now you know how to identify all the off-targets that your CRISPR/Cas9 approach can cause. And we’re sure you’re thinking “Is there any way to avoid those off-targets altogether?”
And the answer is…. almost. But we want to leave you hanging there and take our time to better explain it in the next article of Making the Cut, where we will discuss which strategies we can use to mitigate off-targets!
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







