Scientists Reach New Milestone in Mitochondrial Base Editing: TALEDs Induce A-to-G Substitutions in Mitochondrial DNA
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Not so long ago, editing the mitochondrial genome to cure mitochondrial diseases seemed impossible. Then, a series of key breakthroughs changed the game completely. The first was the development of a CRISPR-free base-editing system for mtDNA by David Liu’s team at Harvard. The second was the adaptation of the base-editing system for inducing C-to-T substitutions in mtDNA in mice, by Jin-Soo Kim’s team at the Centre for Genomics at the Republic of Korea’s Institute for Basic Science.
The Kim lab has now overcome the next major step for therapeutic mtDNA editing: inducing A to G substitutions in mtDNA. The study, published in Cell, describes and compares several different systems for inducing A-to-G edits in mtDNA, using transcription activator-like effector (TALE) proteins fused to bacterial deaminases (TALEDs).
Combined with their previous C-to-T editing system, the possibility to now also perform A-to-G substitutions enables the theoretical correction of around 83% of pathogenic single nucleotide mutations in the mitochondrial genome. The implications of the study extend beyond mitochondrial disease, as Kim remarks:
»Mitochondrial genetic variations are associated with cancer, diabetes, and age-related disorders. TALEDs can be a broadly useful tool for studying the relationship between these variations and diseases.«
TALE proteins and bacterial toxins for editing mitochondrial DNA
Kim’s lab was the first to adapt base-editing systems to edit the mitochondrial genome in mice, inducing pathogenic mutations in the ND5 gene via C-to-T substitution. They did so using DddAtox, a bacterial toxin that can catalyse the substitution of cytosine for uracil. DddAtox acts on double-stranded DNA (dsDNA) and was originally used in the cytosine mitochondrial base-editing systems (DdCBEs) developed by the Liu lab.
Using their own genome-scale library of DNA-binding transcription activator-like effector (TALE) proteins, Kim and his team created a system for inducing C-to-T edits by fusing the TALEs to split DddAtox halves and a uracil glycosylase inhibitor (UGI). In this system, the TALE acts similarly to a CRISPR sgRNA by directing the DddAtox enzyme to a target sequence.
»TALE proteins are composed of highly homologous repeat units of 33-35 amino acid residues, which can be recombined during cloning. To make things worse, it is time-consuming to clone TALE-encoding plasmids. Many years ago, we developed a golden gate cloning system, which consists of 424 TALE sub-array plasmids, to facilitate cloning of TALE expression plasmids in a single cloning step. We now use essentially the same method to construct TALE-DddAtox (TALED) plasmids,« Kim explains when asked about the role of TALE proteins in mtDNA editing.
Demonstrating C to T base editing in mtDNA was a major breakthrough; other gene editing systems, like CRISPR-Cas9, cannot be used to edit mtDNA (see Fact Box). However, C-to-T substitutions are only theoretically able to correct around 40% of pathogenic mutations in mtDNA.
In order to correct a majority of mtDNA mutations, Kim’s team needed to develop a system that could induce A-to-G substitutions. In the current study, they sought to do so by using TadA8e, a variant of the TadA deoxyadenosine deaminase identified in E. coli.
»TadA is an RNA-specific adenine deaminase. The Liu lab engineered TadA, by directed evolution, to act on single-stranded DNA, which is fused to the CRISPR-Cas9 nickase to create adenine base editors (ABEs). This is an amazing achievement because there are no known naturally-occurring adenine deaminases working on DNA. TadA-8e is a further-engineered version with enhanced activity and is an essential component in one of the most efficient ABEs,« Kim says.
mtDNA: the final frontier of genome editing
There are 37 mtDNA genes, all of which are vital for mitochondrial function. Unfortunately, mtDNA has a higher mutation rate than its larger nuclear counterpart; 1 in 5,000 people inherit pathogenic mtDNA mutations. Currently, there are 95 clinically confirmed mtDNA mutations that cause mitochondrial disease and 90 of these are point mutations.
Several factors have historically prevented scientists from manipulating the mitochondrial genome. Firstly, the mitochondrion is double-membraned, making delivery of any editing components a key challenge; earlier attempts to edit mtDNA using CRISPR-Cas9 failed to deliver the necessary single guide RNA into the mitochondrion. Secondly, limited recombination means that double-stranded breaks are not efficiently repaired by either non-homologous end joining (NHEJ) or homology directed repair (HDR) mechanisms. As an additional difficulty, the mitochondrial genome is polyploid, i.e., there are many copies within each cell, causing any successful editing to likely result in heteroplasmy, which may not be sufficient to alleviate disease.
The recent success in base editing in mtDNA may offer alternatives to the oft-debated ‘three-parent baby’ dilemma faced by those who suffer from mitochondrial disease. The controversial mitochondrial replacement in vitro fertilisation procedure involves removing the disease-carrying mother’s mitochondria from the egg and replacing them with mitochondria from a healthy donor, either before of after fertilisation. While the nuclear DNA of the mother is retained, the resulting embryo carries mtDNA from the donor, resulting in three biological parents. While therapeutic gene editing comes with its own raft of concerns, base editing can potentially correct the pathogenic mutations without the need for mitochondrial replacement therapy.
First steps: inducing A-to-G edits in mtDNA
The first step was to determine if fusing TALE proteins to TadA8e and a mitochondrial targeting sequence (MTS) could induce A-to-G substitutions in mtDNA. Kim and the team used TALEs designed to bind the ND1 and ND4 mitochondrial genes, mutations in which cause several mitochondrial diseases.
Although the fusion proteins were able to induce A-to-G substitutions in the ND1 and ND4 genes in human embryonic kidney cells (HEK293T), they had very low observed activity − less than 1.2%. It was a start, however, and Kim was determined to continue with the study.
The next challenge was to increase the base-editing efficiency, for which Kim and the team decided to combine TadA8e with the DddAtox enzyme they had used for inducing C-to-T substitutions. They theorised that because DddAtox acts on dsDNA, it might make the DNA more accessible to TadA8e, which works on ssDNA. However, there is a catch to using DddAtox safely, as Kim explains:
»DddAtox is a cytosine deaminase acting on dsDNA. As a result, Dddtox is cytotoxic. In fact, it is an inter-bacterial toxin. To avoid cytotoxicity, it has to be split or inactivated by site-directed mutagenesis.«
Using a pair of TALE arrays, the two split halves are brought back together at the target site for editing. With this TALE deaminase (TALED) method, the team were able to increase the frequency of A-to-G substitutions, reaching 19% in the HEK293T cells (Figure 1).
While this result was very encouraging, the problem with the use of TadA8e together with DddAtox is that the system induces C-to-T substitutions as well as the desired A to G edits.
The solution was to remove the UGI; UGI-free TALE deaminase pairs targeted to ND1 were able to induce A-to-G substitutions with up to 49% efficiency, without creating C-to-T substitutions or unwanted indels. Interestingly, the position at which the DddAtox enzyme is split was also shown to affect the efficiency of the editing.
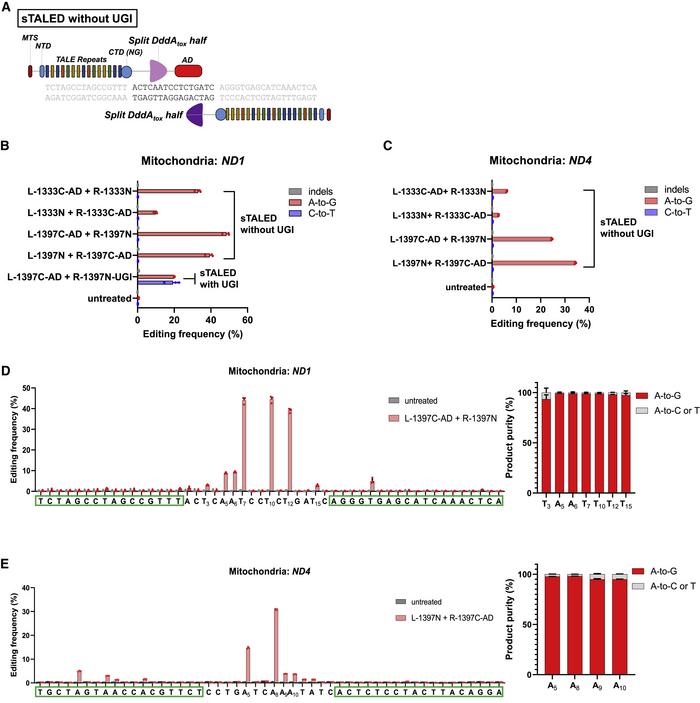
Finding the ideal TALED system through rigorous comparison
While they had increased A-to-G substitution efficiencies considerably, Kim and his team still wanted to perfect the system. One key consideration was whether the split DddAtox was better for editing than a full-length variant with a mutation in the active site to prevent cytotoxicity.
Using the full-length DddAtox, the team also wanted to perform a comparison of monomeric and dimeric TALEDs. Monomeric TALEDs were constructed with a single TALE array fused to TadA8e and DddAtox, and dimeric TALEDs contained two neighbouring strand-specific TALE arrays, with one fused to TadA8e and the other fused to DddAtox.
»We wanted to determine whether DddAtox functions in cis (as in a monomer) or in trans (as in a dimer) with TadA-8e. Both systems worked fine. Monomeric forms are easier to deliver in vivo because you will need a single viral vector, but they tend to induce unwanted bystander edits far from the TALE-binding site. Dimers are less likely to induce bystander edits,« Kim remarks.
For comparison purposes, they targeted all three systems – split (sTALED), monomeric (mTALED), and dimeric (dTALED) – to 11 additional mtDNA gene targets in HEK293T cells. While all were successful, the split TALED system (sTALED) worked the best, achieving A-to-G substitutions at up to 27% efficiency across all 12 sites.
Kim and his team also wanted to explore and define the ‘editing window’ of these three TALED systems. Adenine residues downstream of the TALE-binding site were not efficiently edited, with the best results roughly in the middle of the binding site for each system. The authors then worked to create a shifting editing window to expand the potential of therapeutic editing.
»We were able to shift editing positions by making a series of overlapping TALE protein-linked deaminases. As expected, TALEDs induced A-to-G edits several nucleotides downstream of the TALE-binding site,« Kim comments.
Ensuring safety and moving forward with therapeutic mtDNA editing
After comparing editing efficiencies of the three systems at a range of targets, there was still more work to be done in terms of safety. Kim and the team further tested the system in three other human cell lines, and isolated single cell clonal populations to determine if the edits are maintained over longer time periods.
Kim observes that heteroplasmy, where more than one mitochondrial genomic variant exists within a single cell, remains an issue.
»Heteroplasmy may not be enough to show phenotypic changes. In our paper, we were able to obtain cell lines with 99% edits after drug selection. We will need to develop methods for inducing homoplasmy without drug selection,« Kim says.
They also examined the effects of TALED-mediated mtDNA editing on mitochondrial functions to ensure that editing was not cytotoxic and did not cause instability or dysfunction in mtDNA. To assess off-target editing of all three systems, the team performed additional sequencing of the entire mitochondrial genomes in the edited cells, and also checked to make sure that nuclear DNA was not affected by the system.
»Off-target mutations may cause side effects and must be avoided as much as possible. We are now working on improving specificity of DdCBEs and TALEDs to avoid off-target mutations,« Kim emphasises.
Kim’s current work has greatly accelerated the field of mitochondrial gene editing. While DdCBEs (developed by the Liu group) enabled around 40% of the known pathogenic mutations in mtDNA to be corrected, the current study has essentially unlocked another 43%. This means that the majority of pathogenic mtDNA mutations can now theoretically be corrected with TALED systems.
Even without the possibility of direct therapeutic editing, researchers in the field of mitochondrial disease have struggled with the lack of accurate disease models. The ability to generate precise models of mitochondrial disorders with both A-to-G and C-to-T substitutions will be enormously helpful to the broader field of mitochondrial disease research.
Kim’s pioneering work continues; his team are currently working towards demonstrating therapeutic mtDNA editing in animal models, as well as many other exciting research projects using these and similar editing systems.
»We’ve also been developing zinc finger deaminases (ZFDs) for nuclear and mitochondrial base editing and plant-compatible DdCBEs for mitochondrial and chloroplast genome editing. These systems allow us to induce C-to-T edits. A-to-G editing with TALEDs has been a risky project. I am glad that our efforts payed off,« Kim concludes.
Link to the original article in Cell:
Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases.
Rebecca Roberts, Ph.D. is a molecular biologist and science writer/communicator based in Queensland, Australia.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







