Single-stranded DNA Homology-directed Repair Enhances CRISPR-mediated Knock-In Efficiency – New Promise for Mendelian Disorders
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

The use of CRISPR-Cas9 to introduce therapeutic genes into the genome (also referred to as knock-in) has shown promise in the treatment of genetic diseases. In contrast to non-targeted knock-in approaches, CRISPR-mediated homology-directed repair (HDR) allows the precise insertion of genetic sequences at specific genomic sites.
A challenge in using CRISPR-directed HDR to insert therapeutic genes into the genome of primary cells is the low efficiency of transgene insertion, especially when the transgenes are large. The use of high concentrations of double-stranded DNA (dsDNA) with Cas9 target sequences (CTSs) enhances the efficiency of CRISPR-mediated site-specific transgene insertion. However, this approach leads to significant cell death and a low cell yield, hindering the clinical use of engineered cells.
In a recent study, a team of researchers led by Alexander Marson at the Department of Medicine, University of California San Francisco (UCSF), developed a new method for high-yield genome engineering in primary cells using a hybrid ssDNA repair template and small-molecule cocktails. The use of ssDNA with Cas9 target sequences in HDR templates instead of dsDNA reduced toxicity and increased the knock-in efficiency and cell yield. Notably, the use of small-molecule combinations that enhance HDR further increased the knock-in efficiency.
“The high toxicity of CRISPR-mediated site-specific transgene insertion using dsDNA in HDR templates does not allow the clinical translation of this approach,” said Brian Shy, MD, PhD, first co-author of the study and clinical instructor at UCSF. “Our findings show that using hybrid ssDNA repair templates reduces HDR toxicity, improving knock-in efficiency and cell yields,” he added.
Dr Shy also noted that, compared with previous CRISPR-mediated knock-in approaches, this system is more appropriate for clinical applications and could be used to manufacture clinically relevant amounts of engineered T cells and haematopoietic stem cells, which could be infused back into patients. Their findings were published in Nature Biotechnology.
Study rationale: Enhancing the accuracy and efficiency of CRISPR-mediated site-specific knock-in
Compared to semi-random integration of transgenes using viruses or alternative methods, targeted knock-in allows the correction or reprogramming of specific locations in the genome. “Targeted knock-in allows you to be much more precise with the changes you are introducing,” Dr Shy explained. “We can disrupt specific genes, correct pathogenic mutations, or make use of native regulatory elements to improve the function of cells. There are also potential safety benefits because we know the exact location we are modifying and can avoid any spots in the genome we think might be risky.”
The team previously developed a method to enhance the specificity and efficiency of transgene insertion using dsDNA HDR templates containing Cas9 target sequences. The dsDNA HDR templates and Cas9 ribonucleoprotein complexes were co-delivered into primary cells through electroporation. Although the knock-in efficiency increased two- to four-fold, cellular toxicity also increased.
“One challenge with nonviral insertion has been the toxicity of the large DNA templates we use to introduce a new gene. To achieve efficient knock-in, we need to deliver high concentrations of these templates into cells,” Dr Shy noted.
Approach: Using ssDNA repair templates to increase knock-in efficiency and reduce toxicity
To reduce the toxicity of HDR templates, the team replaced dsDNA with ssDNA templates because ssDNA is less toxic than dsDNA (Figure 1). The team also added Cas9-binding sites on each end of the templates to enhance their delivery into the nuclei of primary cells. “Because ssDNA templates in our system included Cas9 recognition sites, Cas9 can bind to the templates and drag them all the way to the nucleus through nuclear localisation signals present on the protein,” Dr Shy explained. The team validated the feasibility of this approach by using various templates, target sites, and types of human blood cells.
Commenting on the novelty of this approach, Dr Shy said: “Given what we knew regarding the limitations of CRISPR-mediated knock-in due to DNA toxicity, we focused on how we could improve the DNA templates through structural and sequence variations. Therefore, we used ssDNA templates containing Cas9-binding sites to promote the interaction with Cas9 proteins. That helped improve efficiency and yield without changing other parts of the system. We believe this approach can be more broadly applied to improve the efficiency of other CRISPR-based technologies.”
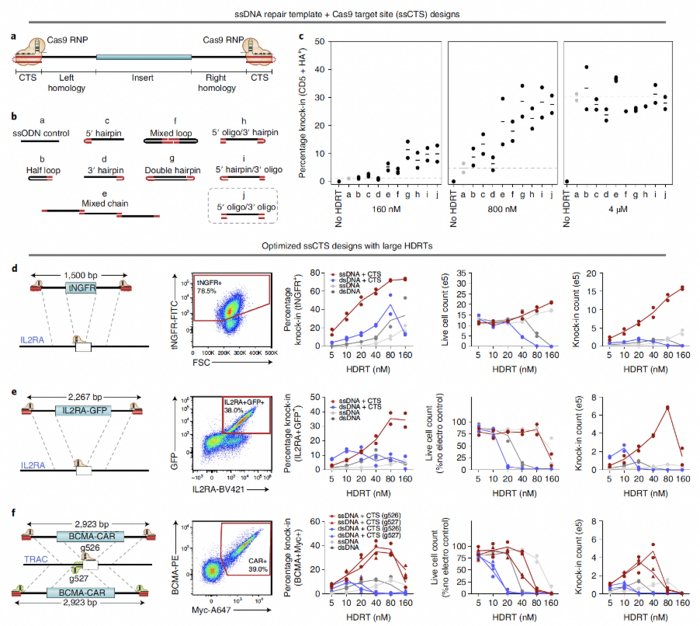
ssDNA templates enhance HDR in primary human cells
The use of ssDNA HDR templates resulted in higher live cell counts compared to when dsDNA HDR templates were used. The absolute live cell counts ranged from 12% to 23% when ssDNA templates were used at concentrations ranging from 5 nM to 160 nM. In comparison, when dsDNA templates were used at the same concentrations as the ssDNA templates, the absolute live cell counts ranged from 0% to 15%. These findings confirm that the toxicity of ssDNA templates is substantially lower than that of dsDNA templates.
Compared with dsDNA templates, ssDNA HDR templates enhanced the knock-in efficiency and yield by two- to three-fold across an array of clinically relevant primary cell types. When ssDNA HDR templates were used, the percentage of knock-in cells was approximately 65% for CD8 T cells; 50–55% for bulk T cells and CD4 T cells; 45% for regulatory T cells; 30% for B, NK, and γδ cells; and 10% for haematopoietic stem cells. However, when dsDNA HDR templates were used, the percentage of knock-in cells ranged from 5% to 25% in the different types of primary cells.
Small-molecule inhibitor cocktails further enhance knock-in efficiency
The DNA-PK inhibitors NU7441 and M3814, the histone deacetylase class I/II inhibitor TSA, the CDC7 inhibitor XL413, and the nonhomologous end-joining (NHEJ) inhibitor Alt-R HDR enhancer have been reported to enhance knock-in efficiency in primary human T cells. Treatment with these inhibitors further enhanced the CRISPR-mediated knock-in efficiency (49% increase for M3814, 46% for XL413, 43% for NU7441, 29% for HDR Enhancer and 16% for TSA) without causing substantial changes in cell viability.
The ability of small-molecule inhibitor cocktails to enhance the knock-in efficiency was also examined. The combination of M3814 and TSA provided the greatest increase in knock-in efficiency, resulting in an approximately 65% increase in the percentage of knock-in cells.
Diagnostic and therapeutic value of CRISPR-mediated knock-in using ssDNA repair templates
Mutations in IL2RA and CTLA4 can cause severe monogenic immune dysregulation disorders. As a proof-of-concept evaluation of the therapeutic value of their method, the team used a ssDNA HDR template with Cas9 recognition sites in combination with an MTX inhibitor to insert the IL2RA or CTLA4 open reading frame (ORF) in primary T cells. This approach provided >80% knock-in efficiency for IL2RA and 70–80% for CTLA4 (Figure 2).
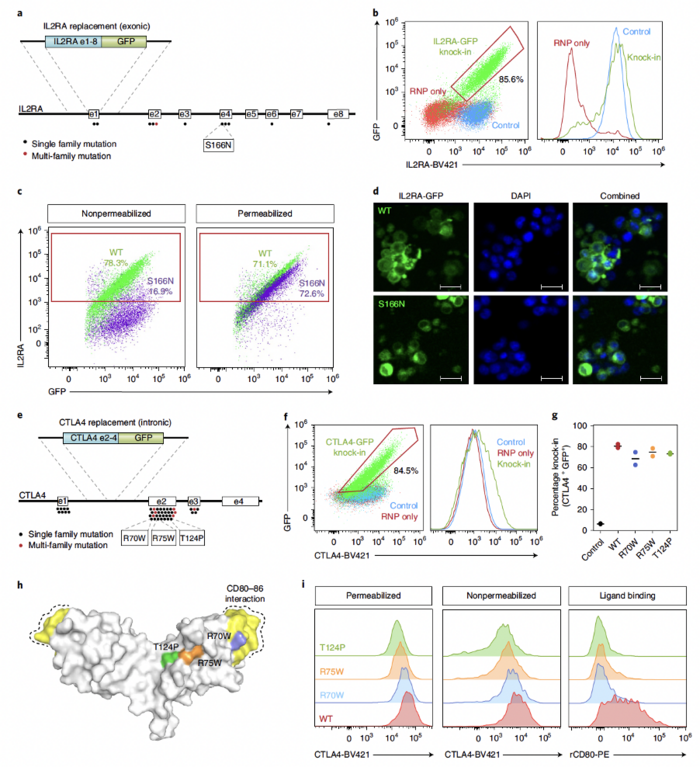
They also used this new approach to express a knock-in construct encoding a disease-causing mutation in exon 4 of IL2RA (S166N). GFP+S166N knock-in cells showed no surface IL2RA, which is consistent with what has been reported in cells from patients carrying this mutation.
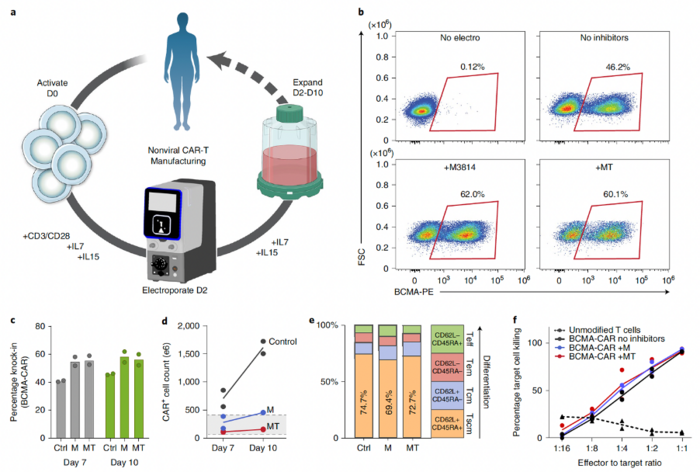
Going one step further in translating their new method into new therapies, the team developed a good manufacturing practice (GMP)-compatible process for manufacturing nonviral chimeric antigen receptor (CAR)-T cells using CRISPR-mediated insertion of a transgene encoding an anti-BCMA-CAR (Figure 3). The average knock-in efficiencies in T cells isolated from two donors were 40.4% on day 7 and 45.8% on day 10, while the yields of CAR+ cells at these time points were >5 × 108 and >1.5 × 109 for both donors.
Challenges
Even though this study confirmed the improved performance of CRISPR-mediated knock-in using hybrid ssDNA repair templates, obtaining large amounts of long ssDNAs remains a challenge.
“To overcome the challenges associated with obtaining large amounts of long ssDNAs, we developed new methods internally and in collaboration with great companies to complete these studies. However, there is still much to be done to simplify and reduce costs with these approaches.”
Looking ahead
The team plans to move this technology toward the clinic at UCSF, hoping that this method will help develop better treatments for patients.
“Now that we have a working system, we are also very interested in further improvements to promote template delivery, reduce toxicity, and combine these approaches with alternative engineering modalities. This will enable even better and more sophisticated cellular therapies in the next generation,” Dr Shy said.
“This is a platform that can be rapidly extended toward other indications, and there are endless opportunities to target cancer, infectious diseases, and many inherited disorders,” he concluded.
Link to the original article in Nature Biotechnology:
Christos Evangelou, Ph.D., is a freelance medical writer and science communications consultant.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







