Targeted Integration With CRISPR
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Welcome back! We hope you had a great Holiday and are ready to learn CRISPR again! Before hanging up our pipettes to enjoy a well-deserved vacation, we mentioned that, on return, we would have introduced you to a new concept: targeted DNA integration!
– Come on, check it if you don’t believe us! –
First off, let us remind you that thus far we mainly talked about using genome editing to disrupt the expression of a gene, a.k.a to knock it out.
In short, this can be achieved using standard CRISPR/Cas9 in its “messier” fashion, by cutting the double-helix of the DNA, or we can be more subtle and use Base Editing. And, in previous episodes, we saw real-world applications of these examples – and even the first ever approved CRISPR therapy, Casgevy –!
However, some of you may wonder if we can do more than messing a gene up. Can we, for example, introduce new information, rather than disrupting the existing one? And what happens in a therapeutic scenario when knocking out a gene does not do any good? Can we still use CRISPR to fix other problems?
That’s great thinking and it makes us happy to see you craving for more.
And who are we to say no?
Targeted integration allows insertion of external DNA in the genome
Let’s imagine that we would like to introduce a new DNA sequence in the genome. And we want to do it at a specific location. With what you learned so far, you know immediately that “targeting a specific location” is a piece of cake for CRISPR/Cas9. But how do we introduce brand-new information in the position cut by the Cas9?
Let’s recall one of our first episodes, where we explained how once the DNA is cut by the Cas9, a whole emergency system is triggered. It is like when a breach in the hull of a submarine or a spaceship is detected: all systems stop, close all the hatches and plug the holes.
Now, in the cell, the NHEJ is the process that plugs the holes quickly, but sometimes it may make mistakes – insertion or deletions - and CRISPR/Cas9 can leverage such mistakes to KO a gene.
On the other hand, HDR takes longer but the repair is flawless. Cause rather than just plugging the hole with whichever material is lying around in the bay, HDR has a proper blueprint guiding the repair process.
This blueprint is the homologous chromosome. Remember in biology class when they told you our cells have two sets of chromosomes, one from mum and one from dad? That’s in part why. Because when damage occurs in one chromosome, which normally happens during DNA replication, our cells can use the other chromosome as the blueprint for repairing, ensuring that the DNA transferred to new cells will be correct.
It won’t surprise you to know that to install new information we need to trick the HDR and use a new blueprint, one we provide!
We can do that by having a look at how HDR uses the blueprint to repair damage. The HDR looks at the damage and then at the blueprint and it tries to find the missing portion by identifying the regions surrounding it, to its left and right. These surrounding regions are called homology arms.
Once these are found, the HDR will use them to align the homology arms with the region surrounding the hole. Whatever DNA portion located between the homology arms of the blueprint will be assumed to be the one that was lost when the double-strand break (DSB) happened. The HDR will diligently repair the DSB following the schematics.
We are sure that by now you got the trick: indeed, we can synthesize a piece of DNA that contains the sequence we want to introduce in the genome, surrounded by homology arms. These latter will correspond to the DSB site generated via CRISPR.
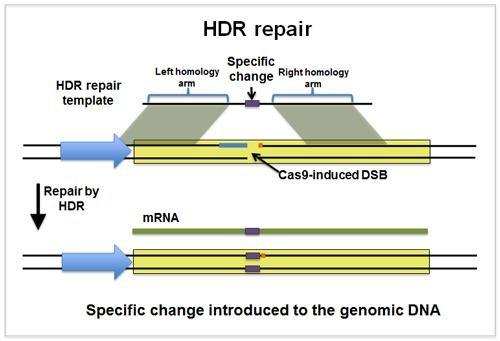
By doing this, we can introduce a DNA sequence of choice at a specific location defined by the CRISPR cut site! This approach is also called a Knock-In or Targeted Integration.
Now let’s see what we can use it for with an example!
Targeted integration to treat SCID caused by RAG1/RAG2 mutations
Imagine that we have a genetic condition where a gene is mutated and the only way to cure the condition is to correct the mutation. We can’t KO gene as we would have done in other instances, we need to repair it.
A good example of a situation like this is the disease called Severe-Combined-Immunodeficiency (SCID) a dreadful name paralleled only by its actual effects. This condition is characterized by an improper development of T cells and B cells, causing dramatic defects in the adaptive immune system with the most severe forms leading to death by the second year of life.

SCID comes in different forms depending on the gene mutated. For instance, in RAG-SCID or Omenn disease. the RAG-1 or RAG-2 genes can be mutated. These genes are quite important cause they are involved in the ability of T cells to recognize different antigens and thereby different threats. Without the activity of these genes, the patient's immune system has some trouble recognizing all the different pathogens (e.g. bacteria, viruses) and even cancer cells the body may encounter. As you can imagine it is like having the immune system “blindfolded”, leaving the person defenseless.
The reason why we bring up RAG-SCID is because the RAG1/RAG2 genes can be mutated at multiple locations in the same patient or across different patients. This means that using a strategy like a Base-editor – which can only change one base at a time – is not a solution.
In such a case, the best way it is to replace the entire gene sequence.
That’s when HDR comes into play!
In fact, numerous research have developed strategies where the healthy copy of the RAG2 is installed in front of the mutated one. The most recent strategy was developed by the group of Ayal Hendel in Bar-Ilan University.
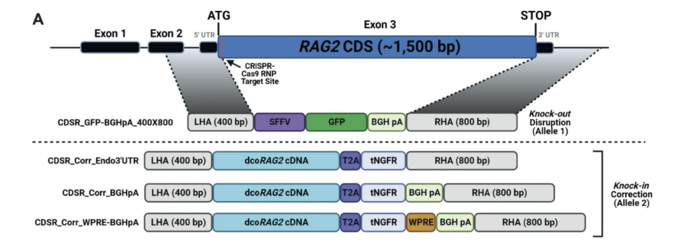
This strategy allows us to cure all the potential mutations in RAG2 at once, cause we are providing the entire correct gene sequence instead of a portion of it. Thus, this strategy can be applied potentially to any RAG-SCID patient independently of where the mutation occurs within the same gene.
This is just an example of what KI can be used for. And there is more to it when talking about basic research applications, but we will stick to the applied side of the matter!
If we left you wanting more, don’t worry! In coming episodes, we will see how instead of curing a disease we may use KI to modify immune cells to fight cancer and even other conditions! We’re getting fancy here!
Till then we hope that you enjoyed our piece and we will see you very soon!
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







