The Dawn of Precision Gene Editing with Programmable Recombinases
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Efficiency, specificity, and safety are essential goals in gene editing. Now, a German team has achieved all three using an innovative approach that transforms non-specific recombinases into highly precise tools for gene editing by custom-designing them to target specific DNA sequences. The process is independent of the cell’s DNA repair pathways and occurs without causing double-strand breaks, thus avoiding potential DNA damage and reducing toxicity.
»While the CRISPR-Cas9 system operates by cutting DNA, leaving the cell’s repair mechanisms to manage the aftermath, our recombinase-based approach circumvents these cellular repair pathways, ensuring high precision. Our comparative studies between the Cas9 and recombinase systems have shown that recombinase edits are remarkably accurate, without any detectable indel formations in the targeted sequences across numerous clones, unlike the CRISPR-edited sites,« says Frank Buchholz. He is a professor and group leader at Technical University Dresden, Germany, where the new technology originated.
Natural recombinases like the Cre-loxP system are of limited relevance in therapeutic applications because of the absence of their target sites in the human genome. Moreover, it is complex to engineer new target sites for recombinases given their combined DNA binding and enzymatic functions within a single molecule, and the process typically takes a couple of months. So, as an alternative to make recombinases more suitable for practical gene editing, it was beneficial to develop a new approach to custom-design their target sites. The solution turned out to be zinc finger domains (ZFDs).
Zinc fingers direct recombinase activity
»Zinc finger domains are modular, with each domain comprising about 30 amino acids and binding to three base pairs. A typical zinc finger protein like Zif268 has three such domains, allowing it to bind to nine base pairs. These domains can be reconfigured to create zinc fingers of various lengths to match the desired target site,« explains Liliya Mukhametzyanova, the first author of a recent paper in Nature Biotechnology where the new gene-editing approach is decribed. She is a former PhD student in Frank Buchholz’s group and now senior scientist at Seamless Therapeutics.
She explains that online tools can suggest potential zinc finger designs for a DNA target sequence and provide the corresponding amino acid sequences to target that site. This approach eliminates the need for complex engineering and is almost as easy as ordering a guide RNA for a CRISPR-Cas9 experiment.
“This method allows for swapping genetic segments, which can be particularly useful for correcting multiple mutations within a single exon. It offers a more cost-effective alternative to creating individual base editors for each mutation associated with a disease”Frank Buchholz
In a first attempt, the team used a flexible linker to fuse a ZFD (Zif268) to either the N- or C-terminal of the Brec1 recombinase. The recombination rates of the fusion proteins were then tested in a plasmid-based assay on a DNA sequence with the lox- and ZFD-binding sites next to each other.
Results revealed that whereas wild-type Brec1 at low expression rates recombined the plasmid with 7% efficiency on average, the C-terminal Brec1–Zif268 fusion achieved 85% recombination efficiency at the same expression rates. Importantly, fusions with other combinations of ZFDs and recombinases showed the same improvements in recombination activity, indicating that the strategy can be transferred to other programmable recombinases and ZFDs.
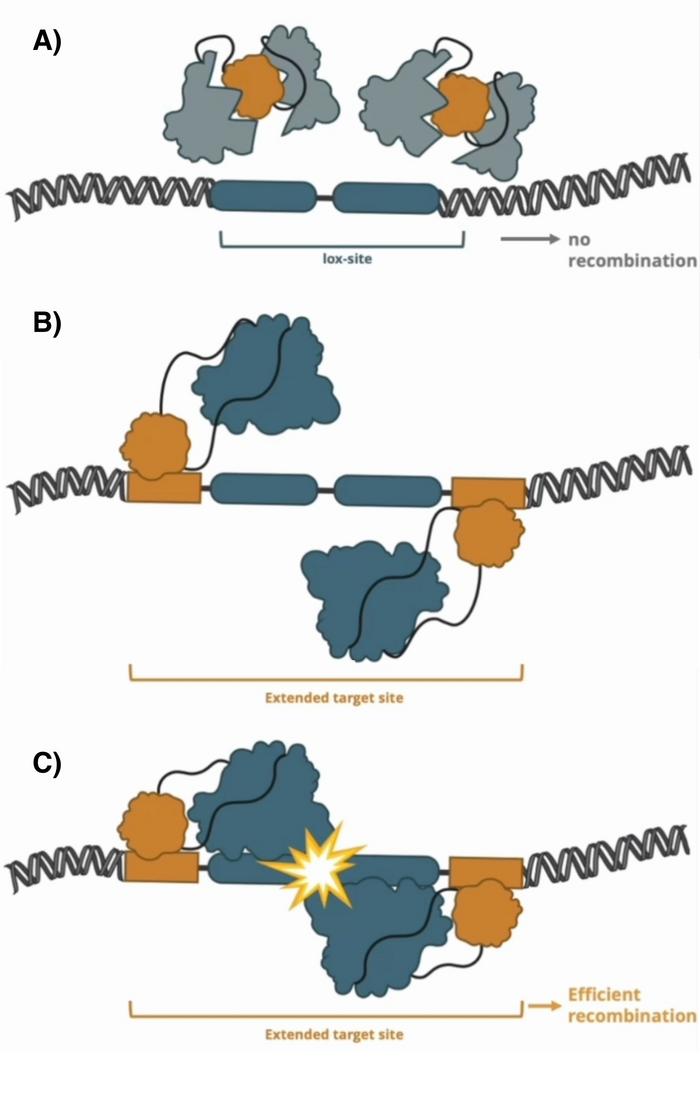
Since it’s much easier to engineer a desired binding specificity to ZFDs than to recombinases, the team speculated that a programmable recombinase-ZFD fusion protein that relied heavily on the ZFD-binding site would be easy to customise and also very efficient. So, they came up with the idea of developing insertional fusions of the ZFD into the recombinase. The insertion initially disrupts the recombinase’s conformation and activity. However, when both the recombinase and the zinc finger bind to their designated sites, the two halves of the recombinase come together and trigger the enzymatic recombinase activity (see Figure 1).
»A pivotal discovery in our research is the ability to inactivate a recombinase enzyme by internally integrating a zinc finger DNA-binding domain, rendering it non-active. However, we can restore its activity conditionally by extending the lox site to include zinc finger binding sites at an optimal distance. When the zinc fingers bind to their specific sites, they activate the recombinase,« explains Frank Buchholz.
Felix Lansing, chief technical officer of Seamless Therapeutics, the company advancing this novel technology into therapeutic application, emphasises that this novel approach significantly enhances activity. Additionally, it may be used as an engineering option to increase specificity by reducing unintended recombination events. Seamless has obtained the exclusive license for the technology and is currently working on building a pipeline of therapeutic candidates. Felix Lansing originally worked with Frank Buchholz and Liliya Mukhametzyanova to develop the new gene-editing method. He says:
»This novel approach allows us to potentially target regions in the genome that we considered before as unspecific loci. With the addition of the ZFD, we can now target these regions as the whole complex recognises a larger stretch of DNA. Together with our designer recombinases, this offers a versatile toolbox, enabling us to blend the strengths of different methodologies to achieve the desired genetic modifications efficiently.«
Recombinase-ZFD fusions correct a haemophilia A-causing inversion
The functionality of the insertional recombinase-ZFD fusions was tested in human HEK293 cells for recombination of a genomic locus. Here, they used the heterodimeric Cre-derived recombinase D7, recently developed for correcting the 140-kb genomic int1h inversion in the F8 gene causing haemophilia A. D7 targets the loxF8 target site in F8, and directed evolution was used to design ZFDs targeting each side of the genomic sequences flanking loxF8.

The ZFDs were then fused internally in D7, and the constructs were transfected into human cells. HEK293T cells carry F8 in the normal orientation, and successful recombination would invert it into the int1h disease orientation. PCR-based assays revealed a 3.8-fold increase in inversion efficiency with the D7-ZFD construct compared to wild-type D7 (see Figure 2).
The approach was also feasible for correcting the haemophilia A-causing inversion. When D7-ZFD mRNA was transfected into patient-derived F8 int1h-hiPSCs, a four-fold increase in inversion efficiency compared to wild-type D7 was observed. A series of experiments further demonstrated that, in contrast to native D7, the D7-ZFD fusion construct did not recombine any potential off-target sites in the human genome.
Recombinases excel in modifying larger DNA segments, one of their primary strengths. However, they can also achieve precise edits at the single nucleotide level, employing techniques like recombinase-mediated cassette exchange.
»This method allows for swapping genetic segments, which can be particularly useful for correcting multiple mutations within a single exon. It offers a more cost-effective alternative to creating individual base editors for each mutation associated with a disease,« Frank Buchholz explains.
Another essential feature for clinical translation of the novel gene-editing approach is the relatively compact nature of recombinases. When coupled with an additional zinc finger domain, their total size amounts to roughly 1,300 base pairs. This compact size allows them to easily fit into adeno-associated virus (AAV) capsids for delivery, especially compared to larger systems like the CRISPR-Cas9 proteins, which typically range between 4,000 and 5,000 base pairs.
Moreover, recombinases operate independently of the cell's DNA repair machinery, which can be variable and often inefficient across different cell types, so they can potentially modify nearly all cells within a targeted tissue.
Patents fuel Seamless’ growth
Though Seamless was founded just over one year ago, the company has garnered significant financial backing, closing a $13 million seed funding round led by venture capital firms Forbion and Wellington Partners, including financial support from the German government. This influx of capital has enabled Seamless to grow its team substantially, from an initial five members to 27 employees based in Germany, all dedicated to advancing their innovative technology.
“While specifics are still under wraps, we’re actively exploring various therapeutic avenues made possible by our programmable enzymes. The diversity of genetic manipulations possible - excision, inversion, exchange, insertion - gives us a broad scope for targeting different diseases”Felix Lansing
» Incorporating zinc fingers has opened up new possibilities for enhancing activity and specificity, providing a flexible tool for addressing various genetic diseases. This adaptability, combined with the inherent safety and predictability of recombinase-based edits - unaffected by the uncertainties of cellular repair mechanisms - underscores the therapeutic potential of our system,« says Felix Lansing.
When asked if he can share any details about the diseases Seamless is focusing on in its research pipeline, Felix Lansing replies:
»While specifics are still under wraps, we’re actively exploring various therapeutic avenues made possible by our programmable enzymes. The diversity of genetic manipulations possible - excision, inversion, exchange, insertion - gives us a broad scope for targeting different diseases.«
Frank Buchholz has been instrumental in developing and protecting the new technology, filing patents early on with an eye toward commercialisation. Seamless's extensive patent portfolio, which it has licensed exclusively, has significantly strengthened its position when engaging with venture capitalists. The company's unique expertise in designing recombinases for specific targets has set it apart, providing a competitive edge that is difficult for others to replicate. The team behind the new gene-editing approach will exploit these advantages and hope to lead the technology into clinical translation.
»We’re actively investigating various applications of the technology in the lab, addressing various severe conditions. This exploration is part of our ongoing research efforts to develop new and valuable genome editing enzymes, broadening the scope of our technological capabilities,« concludes Frank Buchholz.
The article was updated to clarify details of Seamless' funding.
Link to the original article in Nature Biotechnology:
Activation of recombinases at specific DNA loci by zinc-finger domain insertions
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleInterviewNewsHaemophiliaZFN - Zinc finger nucleasesSeamless Therapeutics
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.