Using Engineered Viruses to Deliver CRISPR Ribonucleoproteins: A New Avenue for Therapeutic Gene Editing

Gene editing based on CRISPR technology holds great promise for treating genetic diseases by directly modifying disease-causing mutations. However, in vivo delivery of CRISPR components has proven challenging. Conventional delivery methods based on viral vectors that encode the gene-editing components carry risks of prolonged editing activity and genomic integration.
Administration of ribonucleoprotein (RNP) complexes consisting of Cas endonuclease and single guide RNA (sgRNA) offers many advantages over viral delivery methods, including transient editing activity and lower risk of genotoxicity. However, efficient methods for in vivo RNP delivery are lacking.
In a recent study, researchers at Aarhus University in Denmark developed a new method to deliver CRISPR components with enhanced precision. Specifically, the team, which was led by Professor Jacob Giehm Mikkelsen at Aarhus University’s Department of Biomedicine, engineered lentivirus-derived nanoparticles (LVNPs) that can encapsulate and deliver pre-assembled CRISPR RNPs directly into cells.
»We engineered LVNPs capable of encapsulating Cas9 ribonucleoproteins to overcome issues with the current vehicles for CRISPR components,« noted first author Dr Jakob Haldrup. »Delivery of RNPs using LVNPs led to short-lived but robust gene editing with significantly fewer off-target effects compared to standard RNP nucleofection. Our study helps overcome major delivery hurdles for realising therapeutic gene editing.«
The study was published in Nucleic Acids Research.
Transforming lentiviruses as CRISPR carriers for efficient on-target DNA cleavage
To create LVNPs capable of encapsulating CRISPR RNPs for gene editing, researchers fused Streptococcus pyogenes Cas9 (SpCas9) to two viral proteins, Gag and Pol, which naturally assemble into viral nucleocapsids surrounded by a lipid envelope (Figure 1). This approach allowed for efficient incorporation of SpCas9 during particle assembly. Guide RNAs could be efficiently incorporated into RNPs when overexpressed in the virus-producing cells. Because Cas9 is not expressed by the recipient cells, the RNPs break down within 72 hours.
»We found that fusing Cas9 to the viral core proteins allowed it to be packaged inside LVNPs along with the guide RNA during particle assembly,« explained Dr Haldrup. »This transient delivery system allowed us to control the duration of gene editing activity, unlike viral vectors that cause prolonged Cas9 expression.«
The researchers first demonstrated that LVNPs loaded with SpCas9 RNPs could achieve gene editing in human cells. When SpCas9 was fused to the C-terminus of Pol (viral integrase), LVNPs — which they termed ‘LVNP1.0’ — led to indels in the target gene in up to 40% of recipient cells (Figure 1).
To further optimise the LVNPs, the team fused SpCas9 to the N-terminus of Gag (LVNP2.0). Incorporation of scaffold-modified sgRNAs into LVNPs further increased CRISPR-directed gene disruption, reaching indel frequencies of nearly 100%. The team also refined the stoichiometry of packaging plasmids to increase LVNP production while maintaining high levels of gene disruption.
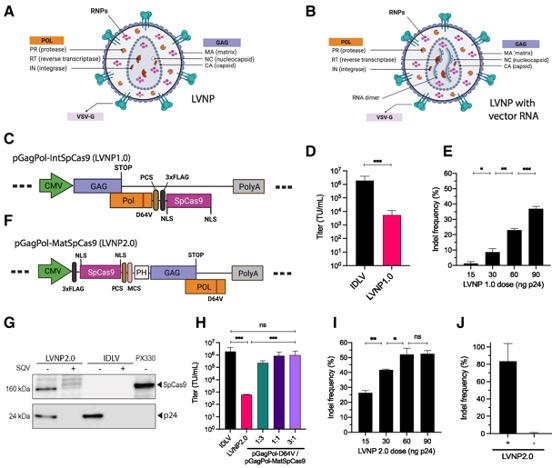
Figure 1. Engineered LVNPs successfully deliver SpCas9 to human cells, leading to gene editing in recipient cells. (A, B) Schematic representation of LVNPs. (C–E) Schematic representation of SpCas9 fused to the C-terminus of Pol, functional titres, and indel frequencies in AFF1 after treatment with increasing doses of LVNPs. (F–J) Schematic representation of SpCas9 fused to the N-terminus of Gag, functional titres, and indel frequencies in AFF1 after treatment with increasing doses of LVNPs. Haldrup et al. Nucleic Acids Research (2023). https://doi.org/10.1093/nar/gkad676.
Lowering the risk of off-target effects
The team found that the editing activity of Cas9 delivered using LVNPs was rapid and short-lived. Indels induced by the delivered RNPs were detectable after 8 hours, and the rate of detectable edits plateaued within 24 hours (Figure 2). Intriguingly, SpCas9 activity decreased to undetectable levels within 4 days of LVNP treatment.
»The short-lived activity gives tighter control over the editing window, which could enhance safety for therapeutic use,« explained Dr Haldrup. »Prolonged genomic activity of conventional viral vectors increases the risk of undesirable genotoxic events.«
One major hurdle in the clinical translation of gene editing is off-target effects due to Cas9-mediated cleavage of DNA at unintended sites that partially match the target sequence. This study demonstrated that RNP delivery using LVNPs resulted in significantly fewer off-target effects than direct delivery of RNPs via nucleofection (off-target indel frequency in mouse hepatocytes: ~70% vs. ~0%). Five-fold higher doses of LVNPs were required to induce detectable off-target effects, whereas nucleofection caused substantial off-target mutagenesis even at low RNP doses (Figure 2).
»We were very surprised and excited to see much higher on-target specificity with LVNP delivery because delivery of RNPs using LVNPs and nucleofection are fairly similar,« said Dr Haldrup. »The lower abundance and shorter activity of RNPs delivered using LVNPs appears to restrict off-target editing while still allowing sufficient accumulation at on-target sites.«
According to the authors, the rarity of off-target effects might be explained by the intracellular trafficking of low-abundant RNPs, allowing accurate delivery to intended sequences. This enhanced precision and programmable activity could significantly improve the safety of therapeutic gene editing using CRISPR.
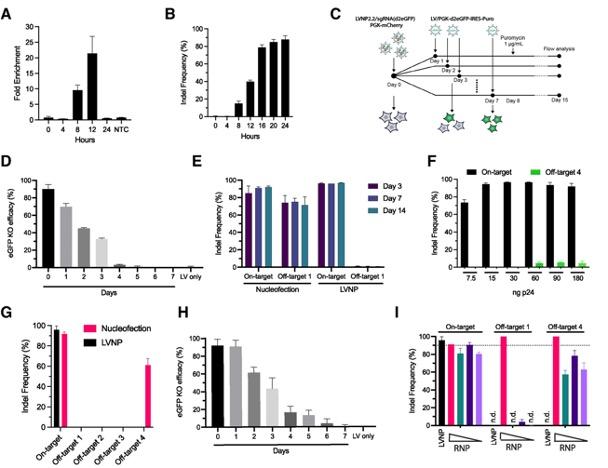
Figure 2. Short-term DNA cleavage activity and reduced off-target effects by LVNP-directed genome editing. (A, B) Double-stranded DNA breaks and indel frequency in Pcsk9 over 24 hours. (C, D, H) RNP kinetics in HEK293T cells by dual fluorescence assay following LVNP treatment (D) or nucleofection (H). (E–G) On-target (Pcsk9) and off-target effects in murine AML12 hepatocytes following RNP delivery using nucleofection or LVNPs. (I) Indel frequencies in AML12 cells after treatment with LVNPs or decreasing amounts of Cas9/sgRNA RNP nucleofection. Haldrup et al. Nucleic Acids Research (2023). https://doi.org/10.1093/nar/gkad676.
Delivering advanced editing systems
The application of LVNPs also extended to the delivery of newer CRISPR editing systems, including base editors and prime editors. These newer editing proteins comprise Cas9 nickase fused to other enzymes that enable precise DNA changes without cutting both DNA strands. The large size of base editors and prime editors makes their delivery very challenging.
LVNPs successfully encapsulated base editor or prime editor RNP complexes and facilitated editing in human cells. Compared to plasmid transfection, LVNP delivery of a base editor resulted in reduced bystander editing at off-target adenines (5%–30% vs. 60%–80%). Prime editing occurred with no detectable indel formation.
Commenting on the versatility of the LVNP delivery approach, Dr Haldrup said: »A major advantage of the LVNP system is the ability to deliver base- and prime editors as RNPs, overcoming size limits of gene delivery vehicles. Being able to deliver base- and prime editors as RNPs provides key advantages for therapeutic use. Their activity is tightly controlled, limiting edit duration, and no genes are transferred into the genome of recipient cells.« Further optimisation is, however, needed to improve prime editing efficacy.
In vivo proof-of-concept
To assess the potential of LVNPs to deliver CRISPR components for therapeutic applications, the team injected LVNPs into the subretinal space of mice to disrupt the Vegfa gene in vivo by delivering Cas9 RNPs (Figure 3).
LVNPs loaded with Cas9 RNPs successfully disrupted the Vegfa gene in retinal pigment epithelium cells, inducing indels in nearly 20% of the Vegfa alleles 4 days after a single LVNP injection. Notably, LVNP injections did not result in detectable off-target effects. These findings provide proof-of-concept evidence to demonstrate the potential use of LVNPs to perform gene editing in vivo for therapeutic applications.
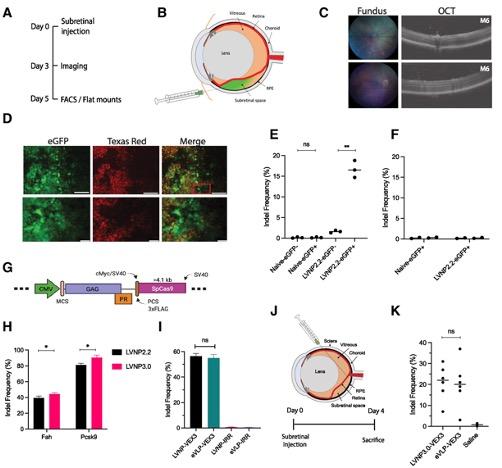
Figure 3. LVNP-directed Vegfa gene disruption in the murine eye. (A–D) LVNP-mediated delivery of CRISPR to edit Vegfa in the murine eye. (E, F) Frequency of indels in Vegfa and at two off-target loci. (G, H) Comparison of the performance of two LVNPs, both carrying SpCas9 and sgRNA targeting Fah and Pcsk9, in Fah reporter cells and AML12 hepatocytes. (I) LVNP-mediated Vegfa indels in transgenic HEK293T cells expressing Vegfa. (J, K) LVNP-mediated Vegfa disruption after injection of LVNP3.0 into the murine eye cup. Haldrup et al. Nucleic Acids Research (2023). https://doi.org/10.1093/nar/gkad676.
Looking ahead
»We’re still early in the translational process, but LVNPs represent a promising carrier for state-of-the-art genome editing toolkits,« noted Dr Haldrup.
According to Dr Haldrup, the next step is to comprehensively characterise the composition of the particles and develop a pipeline for large-scale and GMP-compliant manufacture before testing LVNP delivery in small and large animal models of human disease. Further particle engineering and testing of different administration routes will help advance this method towards future clinical trials.
»This study helps bring the vision of safely editing genes in vivo closer to reality,« he said. “We’re hopeful this technology will eventually help translate gene editing into safe CRISPR therapeutics.”
Link to the original article in Nucleic Acids Research:
Christos Evangelou, PhD, is a freelance medical writer and science communications consultant.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







