Base Editing Slams the Door Shut on HIV

According to recent data from the Joint United Nations Programme on HIV/AIDS (UNAIDS), more than 37 million people were living with HIV, and more than 600,000 had died because of AIDS in 2020 alone.
Although antiretroviral therapy (ART) has dramatically improved the quality of life for HIV patients by suppressing infection and preventing transmission, it is not a curative treatment and requires lifelong administration. Importantly, ART medicaments are effective only if taken regularly. One of the major ART treatment failures is indeed associated with poor adherence of the patient to the therapeutic plan. This can be due to, e.g., medicament side effects, socio-economic situation, and poor patient-physician communication.
New curative treatments are urgently needed, and recent research led by Dr. Mark Osborn at the University of Minnesota Twin Cities suggests that base editing might be the answer. Using this approach, his group simultaneously knocked out the two receptors (CCR5 and CXCR4) that HIV uses to gain entry into cells and start an infection. In already infected patients, this effectively blocks the virus - that is integrated into the patient’s genome - from infecting new immune cells. While this is not a cure, the approach can potentially halt the progression of AIDS, much like ART therapy is currently doing.
Exploring gene editing as a therapy to HIV
Novel therapies based on CRISPR-Cas and other gene-editing technologies are currently being assessed in more than 100 different clinical trials. Moreover, therapeutic strategies based on CRISPR-Cas9 have already yielded positive clinical data in patients affected by sickle cell disease, beta-thalassemia and transthyretin amyloidosis.
Within HIV, the FDA recently approved a CRISPR-Cas9-based candidate to enter a Phase I clinical trial. This candidate, EBT-101, is designed to remove the HIV sequence integrated within the human genome by excising it, thus potentially curing the patient. Beyond CRISPR-Cas, base editing has also shown immense potential to correct genetic diseases in animal models, and the very first base editor, BEAM-101, was recently approved for a clinical trial for sickle cell disease.
Base editing represents an up-and-coming technology in human applications. Furthermore, since base editing does not introduce DSBs, it dramatically reduces the risks of gross chromosomal aberrations (e.g., translocations and chromosomal fusions) that are often associated with traditional CRISPR-Cas9 gene editing. Therefore, base editing holds the promise to deliver both efficient and safe precision therapeutics.
Realising the urgent need for new approaches to HIV therapy, Mark Osborn joined David Liu at the Broad Institute of Harvard and MIT in a multicentre research project to apply base editing to disrupt how HIV infects cells. Their findings were recently published in Molecular Therapy, and we interviewed Mark Osborn to hear more about the study.
Abrogating viral infection by shutting the doors to the immune system
HIV attacks the immune system predominantly through the infection of CD4+ T cells. The virus gains access to these cells via the CCR5 cell surface receptor. Around 10% of the European population carries a 32-nucleotide deletion (Δ32) in the CCR5 gene, providing innate immunity against the CCR5-tropic virus. Therefore, allogenic transplantation of haematopoietic stem cells (HSCs) carrying the deletion is an attractive therapeutic strategy, and this was pursued in the case of Timothy R. Brown, also known as the Berlin Patient, who was the first person ever to be cured of HIV. However, the limited availability of such cells and matching donors represents a considerable bottleneck to this approach.
The story of the Berlin Patient has inspired precision approaches aimed at editing the patients’ HSCs ex vivo to delete CCR5 and then reinject them back into the patient. However, even if CCR5 represents the main entry point of the virus, as the infection progresses, the virus may also exploit the CXCR4 receptor. Keeping this in mind, Osborn's team rationalised that disruption of these two receptors should prevent viral entry to the cells and thus prevent infection. Given the risks associated with introducing two DSBs with CRISPR-Cas9 - one for CCR5 disruption and one for CXCR4 disruption - they turned to base editing, as Mark Osborn explains:
»Particularly for multiplex genome editing, the potential of genome translocation upon introducing two double-strand breaks drove us to ask the question whether we could achieve the same outcome - CCR5 and CXCR4 knockout - using base editors instead.«
Base editors simultaneously disrupt CCR5 and CXCR4 in CD4+ T cells
In an attempt to disrupt both the CCR5 and CXRC4 receptors, Dr Friederike Knipping from Mark Osborn’s lab, who lead the experimental part of the study and is first author on the recent publication, devised a strategy employing both adenine base editors (ABEs) and cytidine base editors (CBEs).
ABEs were used to disrupt the start codon (ATG to GTG or ACG), while CBEs were used to install premature stop codons (TGA, TAG or TAA) in the targeted sequence (Figure 1).
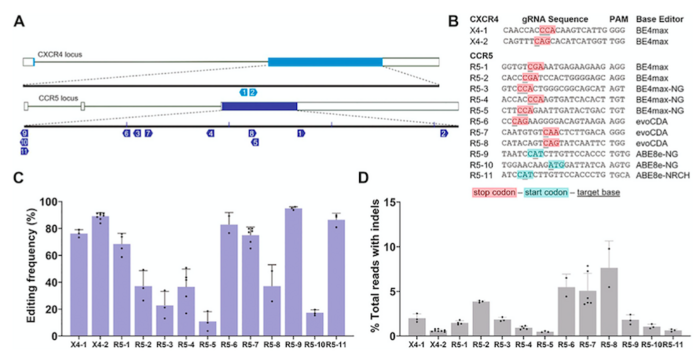
The first step was to test a selection of single-guide RNAs (sgRNAs) and different base editors (BE4max, evoCDA and ABE8e) in vitro in T cells derived from healthy patients. Editing outcomes were analysed via next-generation sequencing, and the combinations were evaluated on the basis of undesired insertions or deletions. Combinations that achieved between 80%-90% of precise editing while minimising DSBs to below 5% of indels were selected. With this set of potent yet safe base editors and sgRNAs, the researchers moved on.
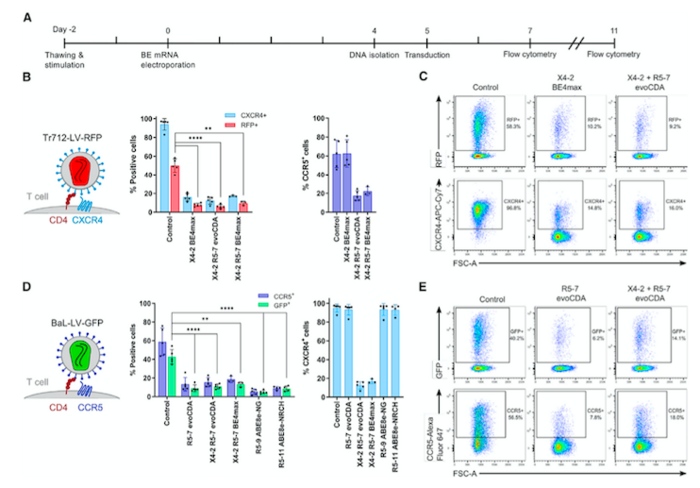
The next step was to interrogate the possibility to simultaneously abolish the expression of CXCR4 and CCR5 in human CD4+ T cells in vitro. As surface membrane receptors, their presence can be easily tracked by staining with specific antibodies and analysing the cells via flow cytometry. This facilitates detection of the fluorescent signal emitted by the antibodies bound specifically to CCR5 and CXCR4.
Upon simultaneous delivery of the best base-editor platform - evoCDA or BE4max - with the most efficient sgRNAs using electroporation of mRNA, a substantial loss in the signal corresponding to antibodies bound to both CCR5 and CXCR4 was observed in the CD4+ T cells. This indicated simultaneous knockout of the two receptors, leading to around 80% CCR5- and CXCR4-negative cells.
»It was extremely exciting because the base editors operated essentially at the same level of standard designer nucleases, so we saw 85%-90% of target protein loss. We were able to target two genes simultaneously and avoid translocation,« says Mark Osborn.
CCR5 and CXCR4 disruption prevents HIV infection
To verify that co-disruption of CCR5 and CXCR4 is a viable strategy to prevent HIV, the team joined forces with scientists from the University of Freiburg to simulate viral infection by using a so-called pseudovirus.
The pseudovirus possesses a modified HIV envelope containing either CCR5- or CXCR4-binding proteins, but it is packaged with a green or red fluorescent protein instead of the viral genomic material. These features make it possible to perform experiments that mimic the infection process without the safety risks associated with using fully functional HIV. This assay delivered base editors and sgRNAs into CD4+ T cells as mRNA via electroporation. Five days later, more than enough to allow the base editors to knockout CCR5 and CXCR4, the edited cells were exposed to the two pseudoviruses.
In this setting, if the pseudovirus successfully infects a cell, it will integrate into the host genome, bringing a transgene that encodes either a green fluorescent protein (GFP, for the CCR5-tropic pseudovirus) or a red fluorescent protein (RFP, for the CXCR4-tropic pseudovirus). The resulting expression of GFP or RFP by infected cells would allow for easy discrimination between cells infected with either of the two pseudoviruses.
The assay indicated that the editing process prevented viral entry into CD4+ T cells. In addition, the experiments showed that the proportion of cells expressing either of the two HIV receptors decreased after base editing. More importantly, while 43% and 51% of unedited cells became infected with the CCR5- or the CXCR4-tropic pseudovirus, respectively, this was only the case for between 5% and 11% of cells edited with various combinations of base editors and sgRNAs (Figure 2).
HSPCs may be the holy grail for base editing
Although this work has demonstrated that co-disruption of CCR5 and CXCRR4 reduces HIV pseudovirus entry into CD4+ T cells, a more attractive cell target would be haematopoietic stem and progenitor cells (HSPCs). These cells can give rise to T cells and other CD4+ cell types, including monocyte-derived macrophages, which are also targets for HIV. Therefore, from a therapeutic standpoint, base editing of HSPCs would likely offer a more extensive level of protection than editing CD4+ T cells alone.
However, this will be more challenging, given that CXCR4 is required for normal physiological function and homing of HSPCs. Unfortunately, strategies to preserve the key functions of CXCR4 - to ensure correct HSPCs development - and at the same time ablate CXCR4-tropic HIV are still ongoing. This means that for now, it is not possible to safely knockout CXCR4 in HSPCs, but Mark Osborn is confident that this challenge can be solved with further work and careful engineering:
»The homing properties of CXCR4 makes it a bit more challenging. The next step is to address technologies that enable robust modification with some maintenance of activity.«
Link to the original article in Molecular Therapy:
Antonio Carusillo PhD is a molecular biologist and freelance science writer based in Germany. He is currently employed at Vector BioPharma.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleInterviewNewsin vivoElectroporationHuman Immunodeficiency Virus Infection, HIVBase editors
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University