Californian Labs Team up to Develop New CRISPR-Tools for COVID-19 Mass Surveillance
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

While the demand for novel gene therapies has been the main driver of the CRISPR-Cas revolution in recent years, the COVID-19 pandemic has catapulted the role of CRISPR within diagnostics.
In May, the US Food and Drug Administration granted emergency use approval for SHERLOCK, a CRISPR-based testing platform for SARS-CoV-2, and the first ever CRISPR tool to be approved for use in humans. Companies around the world are working to bring similar products to market. However, as of yet, none of the newly emerging methods have been scaled up for massive and widespread surveillance of SARS-CoV-2 infection.
»When the pandemic was declared on March 11, I clearly remember having a meeting with Max Wilson and Diego Acosta-Alvear in one of our offices. We saw that the problem was growing completely out of proportion and that there was a huge void. That void was testing. We had the tools, the technical resources, and the molecular biology knowledge. We needed to try to implement some type of testing.«, said Carolina Arias, Assistant Professor at the Department of Molecular, Cellular and Developmental Biology at University of California, Santa Barbara, about the motivation behind her latest work, which has resulted in a new tool for mass COVID-19 surveillance.
Recognising the urgent need for surveillance to find asymptomatic carriers of SARS-CoV-2, Carolina Arias’ lab teamed up with three other labs from the University of California, Santa Barbara. Together, the team (see Fact Box 2) developed a new SARS-CoV-2 surveillance protocol with potential for point-of-care and field deployment. The protocol consists of a cheap RNA isolation method called PEARL (Precipitation Enhanced Analyte RetrievaL) and a new Cas13-based detection assay called CREST – Cas13-based, Rugged Equitable, Scalable Testing.
I recently spoke with Carolina Arias to find out more about the work on PEARL and CREST, the results of which have been shared on the BioRxiv preprint server.
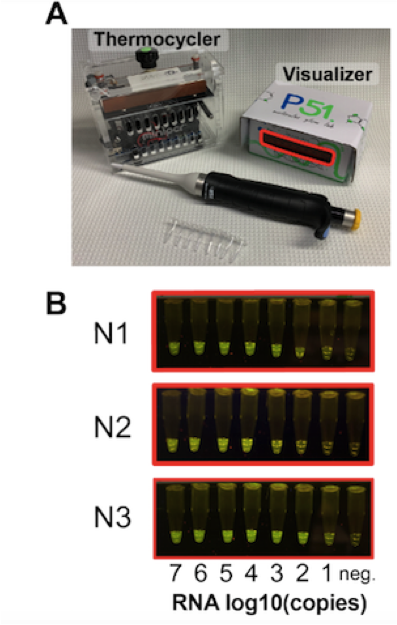
Diagnostics for SARS-CoV-2
Looking at available tests Arias and colleagues realised that all tests had limitations for mass surveillance.
RT-qPCR gold standard methods for SARS-CoV-2 testing rely on the availability of specialised and expensive thermal cyclers.
CRISPR-based strategies such as SHERLOCK, CARMEN (a high-throughput chip-based adaption of SHERLOCK), and DETECTR generally bypass the need for specialised thermal cyclers. Instead, they rely on isothermal recombinase polymerase amplification (RPA), which is much faster and compatible with point-of-care applications. However, upscaling of these CRISPR-based tests for widespread surveillance and monitoring is not trivial because the RPA enzyme is not readily available in large quantities.
The Best of Both Worlds
The team started to explore how they could improve COVID testing to circumvent the limitations of the existing methods.
»We started working with the PCR methods but the reagents were so limiting that we had to find a new way of testing. And that's what motivated us to explore CRISPR assays. We immediately saw that we needed to do something that was not going to be tied up to the reagents that were in short supply at that point.«, explained Carolina Arias.
That is what led to CREST - a unique approach to SARS-CoV-2 detection that combines the specificity of CRISPR and the sensitivity and robustness of PCR with easy-to-access cheap reagents and equipment (see Fact Box 1).
In short, CREST deploys reverse transcription of viral RNA into DNA, PCR amplification, and a single step in vitro transcription and Cas13 detection of specific SARS-CoV-2 sequences.
After optimisation of the Cas13 reaction conditions, the researchers found that CREST could detect as few as 10 copies of a target RNA per microliter of sample, making it as sensitive as standard RT-qPCR methods.
The enzymatic reactions are carried out in a cheap, Bluetooth-enabled, field-ready thermal cycler, and the results are easily seen with a P51 cardboard fluorescent visualiser as shown in Figure 1.
Taking all costs into account (including the expensive instrumentation) CREST fairs out 30-50 times cheaper than established SARS-CoV-2 testing methods.
Fact Box 1: CREST – Cas13-based, Rugged Equitable, Scalable Testing
The researchers developed an in vitro assay to detect the SARS-CoV-2 viral RNA sequences that are included in the U.S. Centers for Disease Control and Prevention’s (CDC) gold standard RT-qPCR test. The assay begins with DNA sequences that represent three regions in the SARS-CoV-2 nucleocapsid gene. These sequences are synthesised chemically as DNA oligonucleotides, and they contain promoter sites for RNA polymerase that permit in vitro transcription to yield SARS-CoV-2 target RNAs.
The in vitro transcribed RNAs are then combined with purified Cas13 endonuclease and gRNAs that programme Cas13 to recognise the viral RNA. A fluorescently labelled RNA reporter molecule is added to the mix and the reaction conditions are optimised for Cas13 endonuclease activity. Once Cas13 recognises the viral sequences it becomes activated, and unleashes non-specific or collateral RNase activity, cleaving the reporter molecule to release a fluorescent signal.
To apply CREST to human testing, RNA is isolated from swab samples, which is then reverse-transcribed to complementary DNA (cDNA) using SARS-CoV-2 target-specific primers. To increase testing sensitivity, the readily available and cheap Taq polymerase is then used to amplify the SARS-CoV-2 cDNA, which is finally transcribed to yield target SARS-CoV-2 target RNAs. These RNAs are then detected by Cas13 and easily visualised with the P51 cardboard fluorescent visualiser.
Bypassing the Need for Commercial RNA Kits
But more optimisation was needed. When the team first developed CREST, they used column-based RNA isolation methods but like many others they quickly saw the limitations of this step.
»RNA isolation was one of the most expensive parts of what we were doing, on a per sample basis. We estimated that it cost around $2 per sample, and we wanted to minimise that. It was also hard to get the column-based kits at times, and we experienced delays.«, said Carolina Arias.
Diego Acosta-Alvear, who runs one of the other three labs, put his team to work to find a safe and cheap solution, and before long they succeeded in developing an easy column-independent way to isolate not only RNA, but also DNA and protein. The method, which they call PEARL - Precipitation Enhanced Analyte RetrievaL - works by first lysing cell membranes and viral envelope proteins in a non-ionic detergent-based solution, and then precipitating out the RNA, DNA or proteins using alcohol.
Optimisation of PEARL allowed the team to detect SARS-CoV-2 in positive swab samples with similar sensitivity to that of commercial RNA isolation kits. PEARL-isolated RNA is fully compatible with the CREST protocol and the team expects it to greatly increase the testing capacity because the reagents used are cheap, found in most labs, and are accessible even in remote areas.
Real-Life SARS-CoV-2 Surveillance
The real test is, of course, how CREST and PEARL perform on real-life samples.
Since sharing their preliminary data on bioRxiv, the researchers have applied CREST to detect SARS-CoV-2 in human oropharyngeal swabs samples and observed 100 % agreement between CREST and established RT-qPCR methods in known positive and negative samples.
The team has also rolled out a pilot surveillance program for asymptomatic volunteers from the UC Santa Barbara campus community in an attempt to monitor asymptomatic carriers of SARS-CoV-2. The results are very promising.
»So far we have tested almost 1,800 asymptomatic individuals, and we found 9 positive cases. The correlation is perfect between the two testing methods.«, explained Carolina Arias during our interview.
The 1,800 individuals tested as part of the surveillance experiment were tested as 2 cohorts; one group of 730 individuals were sampled and tested between the end of May and mid-June, and the remaining individuals were tested from June 23rd to July 2nd. In the first cohort, no positives were identified, but a positive was identified on the first day that testing began for the second cohort. Carolina Arias explains:
»We tested the second cohort about three weeks after parts of Santa Barbara reopened and people could move around freely again. The results of our surveillance testing reflect perfectly what we are seeing in the community in terms of numbers of cases. So we picked up the leading edge of the outbreak. This shows the importance of expanding the capacity further and that is what we are trying to do.«
PEARL and CREST Overcome Hurdles of CRISPR and RT-PCR Methods
Because PEARL and CREST take advantage of widely available and cheap reagents, enzymes and low-cost instruments, the workflow addresses the major hurdles associated with all other SARS-CoV-2 tests and for the first time makes upscaling for massive surveillance possible.
PEARL and CREST can be run with battery operated portable equipment and the workflow has the potential for field use and testing in areas that do not have access to specialised equipment and testing facilities with highly trained personnel. Right now, the team is working to obtain approval from regulatory bodies to roll the assay out in hard-to-reach areas in California in need of testing, and hopes to have this in place by late August.
Link to full pre-print reports:
A Scalable, Easy-to-Deploy, Protocol for Cas13-Based Detection of SARS-CoV-2 Genetic Material.
Fact Box 2: The Researchers
Fromt the Department of Molecular, Cellular and Developmental Biology at University of California, Santa Barbara:
Carolina Arias PhD, Assistant Professor leads a group of researchers on the study of virus-host interactions. The Arias Lab uses cutting edge high-throughput methods such as ribosome profiling and CRISPR-based genetic screens to understand how viruses use host cellular components to translate, fold and modify their proteins.
Kenneth Kosik PhD, Professor leads the The Kosik Lab. The lab explores fundamental biological processes, particularly those related to the brain and its evolution. The research encompasses studies on genes, molecules and cells, as well as large genomic and transcriptional and imaging data set.
Diego Acosta-Alvear, Assistant Professor. The Acosta-Alvear Lab aims to decipher the fundamental mechanisms by which cells respond to stress to preserve homeostasis.
Maxwell Wilson PhD, Assistant Professor leads the Wilson Lab that combines biology, engineering, and physics to understand the cell’s perceptual field.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







