Cas12a: Engineering superior tools for the CRISPR toolbox. Interview: Stefano Stella, Twelve BIO
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
(Interview is condensed and edited for clarity)

- Twelve Bio is a new start-up designing improved CRISPR Cas12a enzymes?
We are developing new Cas12a variants with superior abilities, and we are focusing on two products. One is for genome editing to cure genetic disorders and the other is for diagnostics to identify specific biomarkers for diseases. And here the first system is to identify lung cancer.
- Ok, why Cas12a and not the more commonly used Cas9?
Well, for us it began because we do structural biology research. We want to understand protein-DNA complexes - how a protein recognizes specific DNA targets, how it cuts DNA and so forth. We worked on other systems such as TALENs and meganucleases before the CRISPR era. And then about five years ago Cas12a came out and we thought it was a very interesting protein to study.
- And what makes Cas12a special?
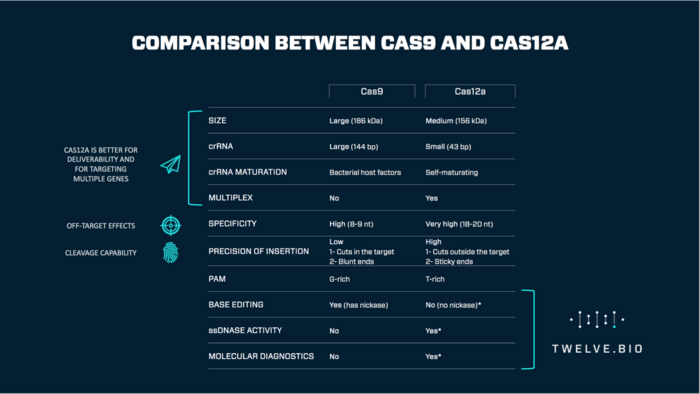
Quite a few things - Cas12a recognize a different PAM than Cas9, the RNA is very small compared to Cas9, but the most interesting for us was a single catalytic site for cutting both strands of the DNA. Cas9 has two catalytic sites - one for each filament of the DNA target - and we were thinking, how does it do that?
- Then You studied its structure?
Yes, we did structural/biophysical and biochemical analysis to understand how the protein was cutting the DNA.
First, we managed to determine the structure of CRISPR-Cas12a after DNA cleavage using X-ray crystallography, and then we used cryogenic electron microscopy (cryo-EM) to study how the protein moves from the moment it recognizes the PAM site until it cuts the DNA. The results were published in Nature and Cell.
3 major check points
- So it was a sort of stop motion picture of the cutting?
Exactly, and thanks to all this information we found three major checkpoints, the protein use to control, when the RNA and the DNA target are hybridized. There is a checkpoint after a few bases, then another between 10-12 bases and the last checkpoint at position 16. So, the protein only becomes competent to cut the DNA after there is activation at all three checkpoints.
- and how is this compared Cas9?
Cas12a seems to be more precise than Cas9 for the recognition of the DNA target.
Biophysical data also from other labs have shown that Cas9 requires only about 8 nucleotides to form a stable complex that is competent for cleavage the target DNA, while Cas12a requires 18 nucleotides to form such a complex. That means Cas12a target sequences are long enough to be unique in large genomes like the human genome.
Also a new activity of Cas12a was discovered in 2018.
- new activity?
It was shown that Cas12a after recognition of the specific DNA target unleashes a very strong single-stranded DNA cleavage activity that can cut a very large number of single-stranded DNA molecules. Thus, the single catalytic center in Cas12a now cuts the two strands of the DNA target as well as single-strand nonspecific DNA.
Engineering superior Cas12a
- ok and then You decided to engineer Cas12a to make it even better?
After identification of the 3 checkpoints, we thought can we change them, and will the protein work differently?
Then by making a combination of changes in this region we managed for the first time to have a Cas12a that has no single-stranded nonspecific activity anymore.
- why is that important?
When you do genome editing the Cas12a, in theory, can cut any single-stranded DNA in a nonspecific manner. So, we think this could be problematic perhaps generating off-targets or other problems.
But now we managed to have a Cas12a without nonspecific activity and still as active as before.
And since we were on the path for modeling this protein, we also managed to obtain a Cas12a that does not cut the target DNA anymore, but only has the single-stranded nonspecific activity. So basically, the other way around.
A very sensitive detection system
- and why is that?
At the end of 2018, the lab of Jennifer Doudna and Feng Zhang managed to link this unspecific activity to a detection system for molecular diagnostics. Thus, when Cas12a recognized the specific DNA sequence the nonspecific single strand activity can produce a detectable signal that indicates the presence of such a sequence.
It is very precise - they managed to use a Cas12 based detection system for two different serotypes of the HPV virus and the difference is only one base.
- and your idea is to improve on this by taking the specific activity away from Cas12a?
Yes. That's exactly the point. We are taking the specific activity away from Cas12a, but not the ability to recognize a specific target. So, we believe that our Cas12a variant can cut more single-stranded DNA molecules compared to the wild type protein, when it is activated by the specific sequence, thus producing a higher detectable signal.
In other words, making our protein more sensitive for molecular diagnostics.
Lung cancer diagnostic tool
- and is there a specific disease You are focusing on?
We are looking at lung cancer by detecting circulating tumor DNA.
We have identified 63 different sequences that have been associated with lung cancer, and we can use the same protein just with different guide RNAs to identify the different sequences. So, our idea is to make an array using these 63 sequences and make an easy point of care system for the identification of lung cancer biomarkers.
- why this particular disease, lung cancer?
Millions of people are affected by this disease and there are no good screening methods for it. Symptoms only occur at later stages when you are already coughing, and a lump is identified by NMR or X-ray. We are hoping an easy system can be very useful for the early detection of lung cancer, which of course has a much better prognosis.
Gene therapy tool
- are You also developing Cas12a as a therapeutic tool?
Indeed, we are testing our variant without single strand nonspecific activity on its ability to edit genes responsible for genetic disorders such as the mutations in the HBB-gene that are the cause of sickle cell anemia.
Furthermore, we are a platform company, and we are also developing new Cas12a variants with new abilities.
- why are you testing for correcting sickle cell anemia disease?
We investigate the HBB gene - sickle cell anemia gene - because it is a very well studied system and there is a lot of information, also from other genome editing systems. We want to have a direct comparison of on- and off-target to compare our protein as fair as possible to all the other systems around.
- ok final question, what is the timeline for you to have a product?
Well, we are very new company, so I think that for other Cas12a variants with new activities compared to the ones we currently have, we work on them all the time, so we could have something soon, but for the diagnostic tools for lung cancer, we think we can have a product may be in 2-3 years.
For genome editing products for curing diseases will be in the long term probably 5-10 years.
Twelve BIO
Twelve BIO was established in September 2019 at the Bio Innovation Institute (BII), thanks to a Bio Accelerator academy grant of 250.000 DKK, by its scientific co-founders Guillermo Montoya and Stefano Stella at the Novo Nordisk Foundation Center for Protein Research at the University of Copenhagen (NNF). It was awarded a pre-seed grant from Novo Seeds of $500.000 (3.5 million DKr).
Twelve BIO is a startup with a technology platform based on CRISPR-Cas12a variants. They specialize in structural biology and protein engineering to design superior Cas12a enzymes for gene editing and diagnostics.
Tags
ArticleInsightInterviewDiagnosticsGene therapyCas12aTwelve Bio
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







