CRISPR-Cas9 base editor caught in the act by Cryo-EM
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

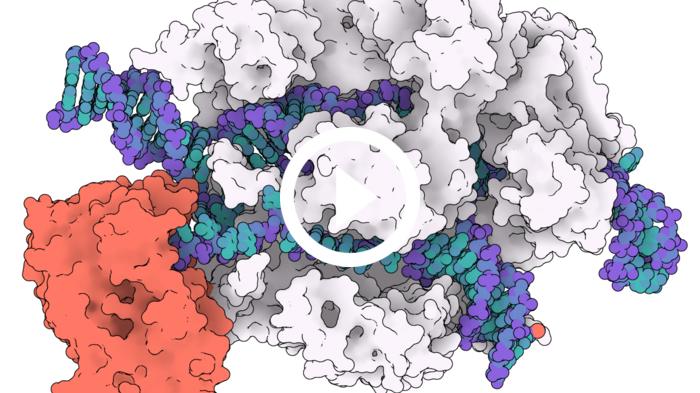
For the first time, researchers have a high-resolution molecular structure of one of the most exciting CRISPR-Cas9 precision genome-editing tools called adenosine base editors (ABE). The highly evolved variant ABE8e with unmatched rapid nucleoside deamination has just been 'caught in the act' by researchers using cryo-EM imaging.
»Our structure of ABE8e is the first structure of a Cas9 fusion. It provides the blueprint not only for how to design more specific base editors but also how to design other types of Cas9 fusions to achieve efficient and precise genome alterations,« says co-first-author Audrone Lapinaite former postdoctoral fellow in Professor Jennifer Doudna's lab at University of California, Berkeley and now Assistant Professor at Arizona State University.
Two main classes of base editors exist: C to T base editors and A to G base editors such as ABE8e, that can collectively make 4 of the 12 possible point mutations and cover about 60% of human pathogenic variants.
»Adenosine base editors are effective tools that have been widely adopted in research science, and they are rapidly moving into the clinic,« says the other first-author Gavin Knott from the Doudna lab and research fellow at Monash University.
Base editors
Base editors are more complex than regular CRISPR-Cas endonucleases. They contain at least two domains: a Cas domain fused to a deaminase domain, and they precisely install targeted point mutations. If existing gene-editing approaches are like 'scissors' that cut the genome, base editors are 'pencils' that enable erasing and rewriting one letter of the genome at a time.
One important advantage to base editors is they don't require double-stranded breaks or donor DNA templates. That means base editors allow precise and efficient gene editing without the risk of unwanted consequences of double-stranded DNA breaks, and without reliance on a template-based homology-directed repair.
Beam Therapeutics co-founded by David Liu is pioneering the use of base editing for human gene-editing therapies.
The potential for therapy is enormous, but little is known about the precise molecular mechanisms that underline base editing.
»While base editors are now widely used to introduce precise changes in organisms ranging from bacteria to plants to mice to primates, we have never before observed the three-dimensional molecular structure of a base editor,« says Professor David Liu, Howard Hughes Medical Institute in Cambridge, who is one of the senior authors.
»This collaborative project with the Doudna lab reveals the beautiful molecular structure of a state-of-the-art highly active base editor, ABE8e, caught in the act of engaging a target DNA site.«
The structure is instrumental according to cryo-EM expert, Professor Guillermo Montoya, Novo Nordisk Foundation Center for Protein Research at University of Copenhagen, Denmark, who helped solve the related CRISPR-Cas12a structure and is engineering new variants for gene editing and diagnostics.
»This is an excellent cryo-EM structure revealing the molecular mechanism of the CRISPR-Cas9 adenine base editor,« says Guillermo Montoya.
»It fills an important gap in the field that has until now limited the rational optimisation of this editing tool. The paper by the Liu and Doudna groups opens the possibility to improve this system and therefore, its therapeutic applications.«
Evolving bacterial proteins into gene editing tools
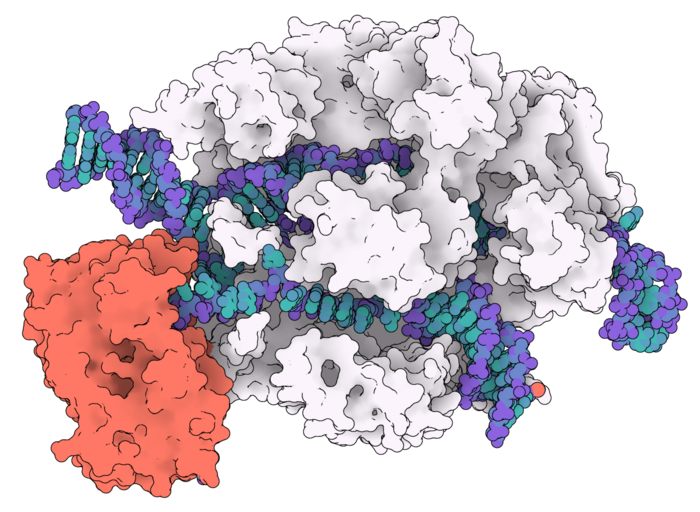
The A to G base editors are fusion proteins that have a deaminase enzyme directly connected to the Cas9 endonuclease - sort of like a DNA scissor carrying a pruning tool. The idea is to take advantage of CRISPR-Cas9 as an incredibly effective 'DNA search engine' that zooms in and locates a specific DNA site, and thereby bring the deaminase enzyme close to the target site.
Once in place, the deaminase prunes adenosine to inosine, which is recognised by the cellular machinery as guanosine.
The DNA repair machinery then repairs the strand, and by using a Cas9 'nickase' that only cuts the unpruned strand the repair machines can be tricked into preferentially using the deaminated strand as a template to build the new DNA double strand.
When the Liu group developed the first A to G base editor in 2017, they took a bacterial deaminase enzyme called TadA (from E. coli) whose active site strictly recognises a rigid tRNA substrate and not DNA. They molecularly evolved the TadA - with 14 substitutions relative to the wild-type TadA - until the enzyme active site specificity relaxed enough to work on DNA presented to it by Cas9.
An extremely rapid base editor
The resulting base editor called ABE7.10 was rapidly applied to many cell types and organisms, and it has also been engineered further into other variants. One such variant is ABE8e which is smaller and has eight more substitutions in the TadA domain. Importantly, ABE8e is incredibly rapid-acting, deaminating DNA at a rate almost 600 fold higher than ABE7.10.
»Since their development, there have been many questions about how these base editors work, and until now, no real experimental data existed to describe the actual mechanism of action,« says Gavin Knott.
Audrone Lapinaite adds: »We wanted to know how the deaminase domain orientates relative to Cas9. Researchers have wondered if it truly matters which CRISPR effector the deaminase is fused to. Gavin and I both had so many discussions about how a tRNA deaminase could evolve to recognise single-stranded DNA,« she says.
Tricking the enzyme to bite
To understand how ABE8e became so rapid and gain a better understanding of base editors in general, the researchers set out to freeze and image ABE8e in action using cryogenic electron microscopy (cryo-EM). The technology won the 2017 Nobel prize in chemistry and is a brilliant technique to reveal the structural details and function of biomolecules in solution. Unlike the traditional method - X-ray crystallography - the proteins do not need to be crystallised, which can be very challenging, but are instead frozen in a thin film, which is then beamed with electrons to create an image.
The extremely fast ABE8e had to be caught at exactly the right moment. It was very challenging but they eventually succeeded, using a modified DNA substrate that mimics the reaction intermediate.
»Basically we tricked the enzyme into biting down on a substrate DNA molecule it couldn't release. It held on to that DNA long enough for us to freeze it in a thin layer of ice for imaging using cryo-electron microscopy,« says Gavin Knott.
The first high-resolution structure
The team produced a 3.2 angstrom (Å) structure that captures the catalytic dynamics of deamination.
»The structure explains several molecular mysteries, including how the mutations that we evolved in adenine base editors unlock their ability to operate on DNA of virtually any sequence,« says David Liu.
»Intriguingly, the structure together with biochemical experiments reveals that these mutations force the target DNA strand to adapt a U-turn shape that mimics the shape of the natural tRNA substrate of the enzyme we started with when we evolved the first adenine base editor in 2017.«
Gavin Knott adds: »We expected that the active site of the deaminase would be wide open so that the DNA that was bound to Cas9 could easily enter the active site. We were surprised to see that the active site was actually narrower,« he says.
The ABE8e had evolved to capture the DNA substrate more effectively, forcing it into the active site so that it can more efficiently modify DNA.
»It helps us understand how base editors broadly work and why the ABE8e variant is so much more efficient than any of the previous generations,« says Gavin Knott.
Off-target effects and how to avoid them
Another aspect is off-target effects - the unintentional editing of bases other than the target base - which is a concern for base editors, especially in therapeutic applications.
The base editors are prone to a particular kind of off-target edit called 'by-stander edits', that occur because the TadA domain will catalyse the deamination of any adenosine groups within reach.
On top of that, transient formation of R-loops in DNA can expose single-stranded DNA that the deaminase can happen to act on at sites utterly unrelated to the target site.
»Due to its fast kinetics, we saw that ABE8e was more prone to perform 'by-stander edits' than the previous versions of ABEs, at least outside of a cellular context. One way of avoiding this type of off-target effect is to reduce the time that ABE8e is bound to the target DNA by using a Cas effector with faster kinetics than SpCas9,« says Audrone Lapinaite.
»One of the most puzzling things we encountered was that ABE8e was able to edit DNA that it was not even targeted to. Based on our activity assays, we speculate that the fast kinetics of ABE8e deaminase domain can capture Cas9 in the state of target search, even before Cas9 forms a guide RNA-DNA heteroduplex.«
She points out that everything was done in highly purified and non-physiological conditions.
»But this ability to edit DNA seemingly randomly needs further investigation, especially in the physiological conditions,« says Audrone Lapinaite.
The blueprint to future designs
The new structure could pave the way for further improvements and molecular engineering.
»This work is another proof of the importance of basic research and the full understanding of a system before thinking about possible applications. The knowledge of the atomic structure of these complexes is a crucial prerequisite to initiate their redesign and improvement for genome editing, without it is like finding a needle in a haystack,« says Guillermo Montoya.
»In the case of the CRISPR-Cas9 adenine base editor, the Liu and Doudna groups have observed and accelerated deamination, which proposes a transient DNA melting during the testing for the target DNA. These details are very important for the future design of base editors.«
Gavin Knott is also excited about future bioengineering.
»One of the major take homes for us was that base editors could be considerably improved if we could find a way to make the deaminase domain more a part of Cas9,« says Gavin Knott.
»Our structure highlights that the deaminase domain is effectively dangling off the side of Cas9. The dream would be to have a base editor whose deaminase domain was so integrated into Cas9 that it was only active when Cas9 bound to DNA, effectively making it allosterically switchable. Our structure provides somewhat of a blueprint for how that might be achieved, and we look forward to seeing where the bioengineers go next in light of our observations.«
Link to original article in Science:
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.