CRISPR-Mediated Correction of ABCA4 Mutations in Stem Cells May Provide Treatment for Incurable Inherited Disease Causing Vision Loss
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Stargardt disease is an inherited macular neurodegenerative disease that causes retinal dystrophy and vision loss. Stargardt disease is caused by mutations in the ABCA4 gene, which encodes a protein involved in the visual cycle and transportation of toxic photoproducts out of the retina.
Currently, there are no treatments for Stargardt disease, and although restoring ABCA4 expression could help prevent or treat Stargardt disease retinal dystrophy, gene replacement therapy has not yet been successful in preventing retinal dystrophy or improving vision.
In a recent study, a team of researchers led by Esther Pomares, PhD, at the Department of Genetics, IMO Foundation (Barcelona, Spain), used the CRISPR-Cas9 system to edit pathogenic ABCA4 mutations in human induced pluripotent stem cells (iPSCs) from two patients with Stargardt disease. This approach led to successful correction of ABCA4 mutations without causing detectable off-target effects.
»For the first time, we have successfully corrected two ABCA4 pathogenic variants in iPSCs derived from patients affected with Stargardt disease. In addition, we have achieved a very high percentage of gene editing, which is important for a potential future therapeutic application,« said Laura Siles Mena, PhD, the first author of the study.
Siles also noted that the use of single-stranded oligodeoxynucleotides (ssODNs) as donor templates for CRISPR-mediated ABCA4 editing ensured permanent restoration of the ABCA4 sequence. She explained that this approach results in higher knock-in efficiency and reduced off-target effects, as well as avoids the use of external sources of DNA. Their findings were published recently in Molecular Therapy Nucleic Acids.
Fact Box: Stargardt disease
Stargardt disease is an inherited condition that causes gradual vision loss in both eyes due to progressive degeneration of the retina, the layer of tissue at the back of the eye that senses light and sends visual signals to the brain. Stargardt disease is the second most prevalent inherited retinal dystrophy (IRD) and is caused by mutations in the ABCA4 gene, which encodes the ATP-binding cassette transporter A4 (ABCA4). Many different mutations in ABCA4 can cause Stargardt disease, and the age of onset and severity of the condition can vary depending on the specific mutation. There are no curative therapies for Stargardt disease, and its management comprises genetic counselling and certain measures to preserve vision, including sun protection, smoking cessation, and avoiding the consumption of high doses of vitamin A.
Study rationale and approach: Using CRISPR-Cas9 to correct ABCA4 mutations
Because all Stargardt disease cases are caused by ABCA4 mutations, gene therapy could be used to restore ABCA4 function by supplying the correct version of the ABCA4 gene. Ongoing clinical trials are investigating the use of gene replacement therapy for the treatment of inherited retinopathies, including Stargardt disease.
Siles explained that the limited success of gene replacement therapy for Stargardt disease is largely attributed to the large size of the ABCA4 gene and the limited cargo capacity of adenoviruses. Therefore, the team decided to test whether gene editing could be a viable alternative for the prevention of retinopathy progression in patients with pathogenic mutations in ABCA4.
The team designed a single-guide RNA (sgRNA) targeting the pathogenic variant they wanted to correct, as well as a single-stranded oligodeoxynucleotide (ssODN) molecule that was used as a repair template for single-nucleotide gene editing.
To achieve CRISPR-Cas9-mediated correction of ABCA4 mutations in patient-derived cells, the team isolated hiPSCs from two unrelated individuals with Stargardt disease and electroporated them with high-fidelity Cas9, mutation-specific sgRNA, and ssODN template.
To reduce the risk of undesired genomic alterations, the team used drugs to enhance ssODN-mediated repair of the DNA double-strand break and inhibit repair through the non-homologous end joining (NHEJ) pathway. The homologous-directed repair (HDR) activator L755507 and the NHEJ inhibitor M3814 were used. Siles explained that although these drugs are not intended for therapeutic use, other drugs that modulate HDR and NHEJ could be used for in vivo application of this approach in humans.
To confirm the successful correction of ABCA4 mutations and analyse potential off-target effects, they established and sequenced single-cell clones of genetically engineered iPSCs. »We sequenced cells using both Sanger sequencing and whole-genome sequencing and found no detectable off-target defects, demonstrating assay precision and effectiveness,« Siles noted.
Moreover, the team analysed the mRNA and protein levels of several stem cell markers in hiPSCs and evaluated their ability to differentiate into the three germ layers to confirm that genetically-engineered hiPSCs retained their pluripotency.
CRISPR-Cas9 using ssODN effectively corrects pathogenic ABCA4 variants
Analysis of hiPSCs from patients with Stargardt disease showed that one patient (Fi22/01) harboured two pathogenic ABCA4 variants, one in each allele (c.4253+4C>T and c.6089G>A, p.Arg2030Gln). The hiPSC line from this patient was termed FRIMOi003-A.
The second patient (Fi15/32, hiPSC line named FRIMOi004-A) had a dinucleotide insertion causing a frameshift in one allele (c.3211_3212insGT) and three missense mutations in the other allele (c.514G>A p.Gly172Ser, c.2023G>A p.Val675Ile, and c.6148G>C p.Val2050Leu). All sgRNAs harbouring patient mutations provided good cleavage efficiency (15%–45%) in patient-derived hiPSCs.
To further enhance the efficiency of CRISPR-Cas9-mediated ABCA4 editing, they designed a ssODN for each variant, which they used in combination with a HDR activator and an NHEJ inhibitor. This approach provided a correction efficiency of 5% for the c.4253+4C>T variant and 11% for the c.3211_3212insGT variant (Figure 1). No genomic alterations (insertions, deletions, indels, or single-nucleotide mutations) were detected in the on-target region, confirming the high precision of ABCA4 editing.
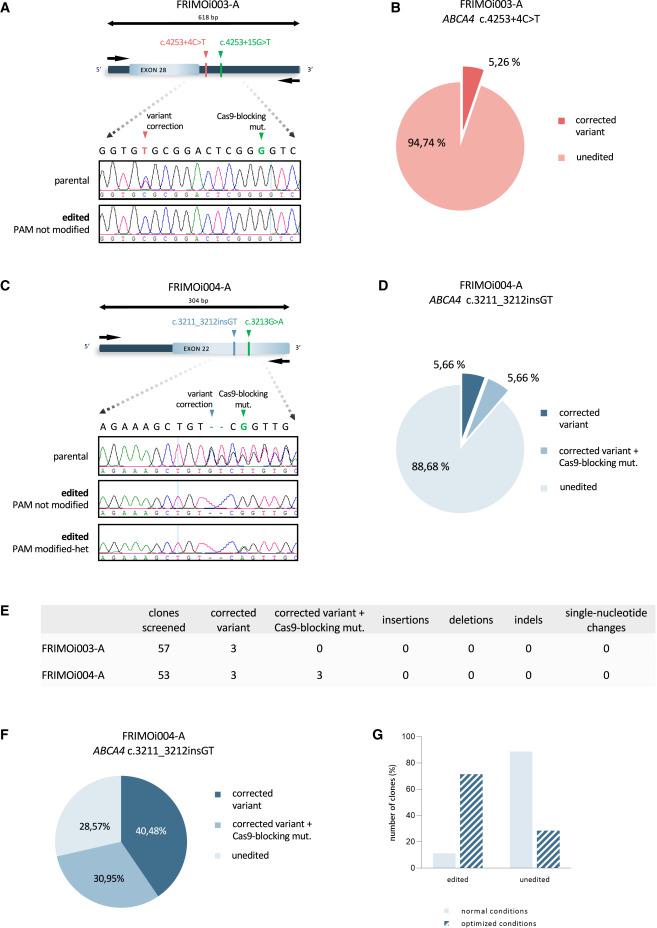
Sanger sequencing also showed that there were no insertions, deletions, or single-nucleotide alterations at off-target sites in any of the 35 genetically-engineered clones screened.
»Our results strongly suggest that specifically designed sgRNAs can effectively discriminate between the two DNA alleles in case of heterozygosity. These analyses also suggest that sgRNAs with more than one mismatch in sequence homology decrease DNA recognition and Cas9-induced DSB,« Siles noted.
Commenting on how many patients are likely to benefit from such an approach considering the number of pathogenic mutations in ABCA4, Siles said: »Stargardt disease accounts for 12% of IRD-related vision loss cases, and more than the 60% of ABCA4 pathogenic variants correspond to missense and nonsense variants while splicing substitutions and small deletions, insertions, duplications, and indels account for approximately 35% of pathogenic mutations. All these pathogenic variants are suitable for ssODN-mediated correction by CRISPR-Cas9 approach performed in our study, providing a potential cure for almost 95% of patients with Stargardt disease.«
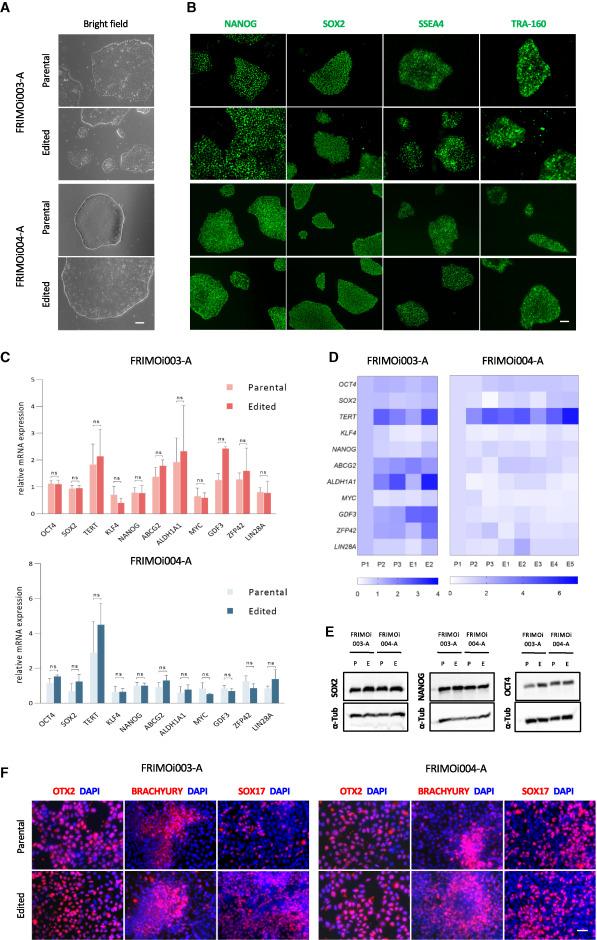
Patient-derived hiPSCs remain pluripotent after CRISPR-Cas9-mediated gene editing
Comparison of hiPSC clones and parental hiPSCs demonstrated that engineered hiPSCs retained their stem cell characteristics, such as colony-like morphology and proliferation (Figure 2). In addition, edited and parental hiPSCs exhibited similar expression levels of the pluripotency markers SOX2, SSEA4, TRA-160, and NANOG. This finding was confirmed at both the mRNA (quantitative PCR) and protein (immunoblotting, immunofluorescence) levels.
The team also analysed the differentiation potential of hiPSCs and found that genetically engineered hiPSCs retained their ability to differentiate into ectodermal, endodermal, and mesodermal lineages despite having been subjected to gene editing. Siles explained that the pluripotency of genetically-engineered hiPSCs is critical to the feasibility of differentiating hiPSCs with restored ABCA4 function into the right cell type for effective reversal of retinal degeneration.
Challenges and future steps
The results of this study demonstrate, for the first time, that IRD-related pathogenic variants can be efficiently and precisely corrected using CRISPR-Cas9; however, this approach is still in the very early stages.
»Further increase in the specificity and precision of gene editing is crucial. Another potentially challenging aspect of developing this approach further is finding a way to avoid re-cutting of the DNA by Cas9 after ABCA4 repair, as multiple rounds of DNA cutting could trigger undesired genomic abnormalities,« she said.
The team also plans to validate the feasibility of this method using in vitro 3D cultures of retinas. »We want to characterise CRISPR-Cas9 effectiveness in organoid models that mimick a potential future in vivo CRISPR delivery strategy for human treatment,« Siles concluded.
Link to the original article in Molecular Therapy Nucleic Acids:
Christos Evangelou, PhD, is a freelance medical writer and science communications consultant.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







