CRISPR Medicine News Interview with Demeetra AgBio CEO, Jack Crawford

(Interview is condensed and edited for clarity)
Where did Demeetra start and what is the company’s focus?
Demeetra AgBio was launched in 2019 as a spinout of Transposagen Biopharmaceuticals (now Hera BioLabs). Our core technology is our proprietary Cas-CLOVER genome-editing tool. This is exclusively licensed to Demeetra and our sister companies Hera BioLabs and Poseida Therapeutics.
At Demeetra, we have rights to use Cas-CLOVER for bioprocessing and agricultural applications. This encompasses editing the chinese hamster ovary (CHO) genome for enhanced biotherapeutic production and editing plants for the production of medically-relevant compounds or to enhance specific crop traits.
Cas-CLOVER technology
How does Cas-CLOVER work?
Cas-CLOVER is a precise hybrid genome-editing system. Similarly to conventional CRISPR-Cas, it functions as an RNA-guided molecular scissors that introduces double-stranded breaks (DSBs) at target sites in genomic DNA (gDNA).
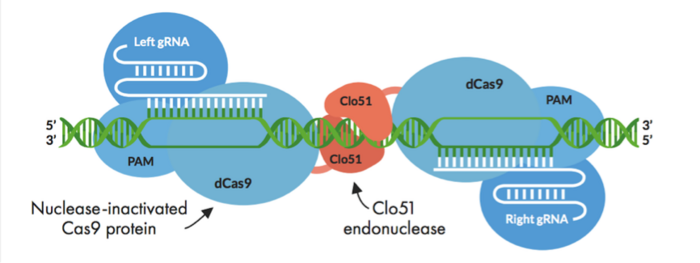
Cas-CLOVER combines the use of an intact but enzymatically inactivated Cas9 endonuclease (dCas9), which is fused to our proprietary dimeric nuclease, Clo51. Unlike conventional CRISPR-Cas, Clo51 is the scissors in Cas-CLOVER and dCas9 merely serves as a fusion protein.
The technology can be used to make deletions, insertions, and site-specific mutations, and it is highly specific. This specificity comes from the fact that Clo51 activity requires dimerisation, where two subunits of Clo51 assemble to form a dimer at the target site, and each subunit is associated with a guide RNA (gRNA).
Are the paired guide RNAs specially designed for Cas-CLOVER?
The gRNAs used for Cas-CLOVER are no different to what is used in conventional CRISPR-Cas, but the requirement for a pair, where each binds to a subunit of the Clo51 dimer, ensures stringent DNA cleavage and makes Cas-CLOVER highly specific.
One gRNA binds to one strand of gDNA, and the other gRNA binds to the other strand. This brings the two dCas9-Clo51 fusion proteins into close proximity leading to Clo51 dimerisation (Figure 1).
As with conventional CRISPR-Cas editing, the gRNAs must bind within a certain distance from a protospacer adjacent motif (PAM) site. This can be challenging, as one needs two find two PAM sites when designing a Cas-CLOVER experiment. However, the spacer region, which is between the binding sites for the two gRNAs, can be 14-40 base pairs (bp) long, which adds flexibility when designing experiments.
Cas-CLOVER vs. CRISPR-Cas
You specifically refer to Cas-CLOVER as being a precise genome-editing tool. What is it that makes it so precise, and what makes Cas-CLOVER distinct from CRISPR-Cas?
Cas-CLOVER editing without the addition of a donor template will lead to non-homologous end joining- (NHEJ) based mutations, which are distinct from CRISPR-Cas NHEJ-based mutations. Cas-CLOVER makes a sticky-ended cut, whereas CRISPR cuts bluntly. We believe that when the cell is provided with a donor template, the sticky ends promote efficient homologous recombination for knock-in and base-pair edits.
Another difference between Cas-CLOVER and CRISPR that we find advantageous is that we can make large deletions with Cas-CLOVER of between 12-40 bp, while CRISPR-Cas generally makes deletions of 1-3 bp. The larger deletions makes it easier for us to identify cells or pools of cells that have the intended deletion – we can easily do this in-house on agarose gels within a few hours instead of relying on sequencing to detect 1-3 bp deletions.
How efficient is Cas-CLOVER and what is the off-target profile like?
For CHO cells, targeted NHEJ knockout efficiency is very high with a typical rate of 70-80%, which is comparable to CRISPR-Cas9. For targeted integration of the gene of interest with selection we see about 90% efficiency, but we have not characterised the efficiency without selection yet.
Since Demeetra is merely using Cas-CLOVER to engineer cellular manufacturing platforms which will never go into patients, we currently do not assess off-target edits unless a client or collaborator asks us to do so. However, our sister company Poseida routinely uses iGuide-seq to profile off-targets after Cas-CLOVER editing in human cells. Here, there are nearly no detectable off-target edits and by far the least when compared with other gene-editing technologies.
How is the Cas-CLOVER editing cargo delivered to cells?
There are probably many ways to deliver the Cas-CLOVER reagents and gRNAs to cells, but we use transfection and nucleofection. What we deliver to cells is a larger cargo that the typical CRISPR-Cas cargo, and so far, we deliver the dCas9-Clo51 as an mRNA. We’ve spent a lot of time optimising the timing for delivering the Cas-CLOVER construct and the gRNAs for each cell type and have developed fully optimised protocols.
Human health applications
How does Demeetra’s work contribute to human health?
In our terms, pharmaceutical bioprocessing is genetically engineering cells that serve as a manufacturing platform for human therapeutics.
We license the Cas-CLOVER technology to companies for strain or cell line improvement. For example, we have clients that use Cas-CLOVER to improve the biologics manufacturing process outside of targeted integration of the gene of interest. This includes, for example, engineering advantageous platforms such as nutritional selection lines based on glutamine synthetase knockout, or cell lines with an enhanced capacity for desired post-translational modifications.
Cas-CLOVER-engineered cell platforms can be based on microbial, plant and other eukaryotic cells and can typically be used over and over for many different products. One can also apply the technology on a case-by-case basis to remove a pesky protein from the downstream isolation processing for example, or to remove a protein whose production simply uses energy that leads to reduced yield of the desired product.
Demeetra's pipeline
So does Demeetra have a pipeline of its own?
Yes, we do. Demeetra is developing CHO cell lines that will be ultra-high yield expressors for undisclosed bio-similar products that are coming off patent or are already off-patent. We are also using our gene-editing platform to identify so-called “safe-harbour” sites in suspension CHO cells. These are genomic locations that permit high and consistent expression of transgenic proteins of interest without disrupting adjacent genes and without detrimental effects to the cell line. Following in depth optimisation of Cas-CLOVER targeting, we expect that targeting at some of our unique safe harbour sites, combined with glutamine synthetase selection, will yield close to 100% efficiencies, which will reduce the timeline and increase quality.
Besides our work in CHO cells, we are also working towards an investigational new drug (IND) application for our lead therapeutic candidate. Here, we are using Cas-CLOVER to engineer plants and yeast strain engineering for optimised cannabinoid production for therapeutic use. We have built very robust internal manufacturing and processing using the cannabis plant where we routinely get 20% yield in the plant and isolate to 99.9% purity on kilogram scale.
And what about therapeutic editing of the human genome with Cas-CLOVER?
All of Demeetra’s human health applications involve editing the genome of cells – human cells, animal cells, plant cells and microorganisms, e.g., yeast cells. We do not work towards editing human genomes in human beings for therapeutic development, but this is possible and is being explored by our sister company Poseida Therapeutics who are approaching clinical development with an allogeneic Cas-CLOVER-edited CAR T product.
Untapped therapeutic potential
Do you see other obvious therapeutic areas where it would be relevant to edit plants to produce either known compounds or derivatives?
Yes, many of the 120+ cannabinoids already produced naturally in cannabis and not produced at levels sufficient for R&D and commercial manufacturing. There are also significant opportunities to enhance natural cannabinoids for therapeutic purposes by using gene editing in combination with synthetic biotechnology. Between editing in tobacco, yeast and eventually cannabis we are working towards building a platform of virtually unlimited bioproduct candidates.
Tobacco is not normally on the top of ones mind when it comes to human health but we use tobacco as a bioprocessing platform and we also have a harm reduction programme using Cas-CLOVER. By eliminating several genes we can remove most of the carcinogens and also modify the nicotine content towards transitioning to reduced harm nicotine products such as chewing gums.
What can we expect from Demeetra in the near future?
Our lead therapeutic candidate is a very specific cannabinoid in a specific formulation to treat a very specific disease and we hope to submit an IND application to the FDA early next year. We envision pursuing regulatory approval for this as a pharmaceutical product.
Tags
ArticleInterviewNewsCARBONin vivoNon-viralDemeetra AgBioCas-CLOVER
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University