CRISPRroots Incorporates RNA-seq to Evaluate Gene Editing Outcomes

After a CRISPR gene-editing experiment, researchers typically sequence both the target locus to verify that it was successfully edited and a few predicted off-target sites to examine if unintended editing occurred. In addition, researchers can extend this work by including RNA sequencing (RNA-seq) to study the transcriptional consequences of the gene-editing events. However, there are multiple potential pitfalls to watch out for.
»If your RNA-seq analyses reveal some genes that are differentially expressed in your control and experimental cell line, you will typically hope that this is the result of CRISPR editing of the target gene. But in reality, it might also be the result of off-target effects or even events unrelated to CRISPR gene editing,« says Jan Gorodkin. He is a bioinformatician and professor at the Center for non-coding RNA in Technology and Health, University of Copenhagen, Denmark and co-senior author of a paper published last week in Nucleic Acids Research.
The paper describes a new software tool, CRISPRroots, that helps researchers better exploit the power of RNA-seq data when evaluating potential off-targets and verifying on-target editing outcomes of their CRISPR experiments. The method compares gene expression and sequence variants obtained from RNA-seq of edited cells and corresponding isogenic controls and combines this information with predicted gRNA-binding properties.
CRISPRroots incorporates three sources of information
»We have three information sources that serve as input for CRISPRroots,« explains Stefan Seemann, the other co-senior author of the paper, who is a computer scientist and bioinformatician working in Gorodkin’s group as an associate professor. Seemann continues his explanation:
»The first source of information comes from an off-target prediction tool that looks for sites where the gRNA might bind based on sequence complimentarities and binding energies. The two other information sources are based on RNA-seq analyses and include sequence variation in the transcriptome and differential gene expression in the edited and non-edited cell lines. In CRISPRroots, we combine all these three data sources and aim to get as much relevant information as possible out of it.«
While other researchers routinely use off-target prediction tools, very few have so far included more than one of the two RNA-seq analyses in their approaches. One reason for this is that the results from RNA-seq analyses can be challenging to interpret.
After a successful CRISPR gene-editing experiment, gene expression variations will include partial or complete loss or modification of the target gene transcript. However, this altered expression of the target gene will typically affect the expression of additional genes because most genes are regulated by and regulate other genes. Consequently, any change in the expression of one gene will initiate a cascade of expression variations in as many as hundreds of genes. The same applies to unintended editing of potential off-target genes, which will cause altered transcription of several genes unrelated to the target gene.
RNA-seq might also reveal alterations in gene expression that are not related to CRISPR gene editing. For example, random mutations in either the experimental or control cell line can result in transcript variations in the two cell lines. Moreover, transcription is an error-prone process that can lead to nucleotide variations between individual RNAs from the same gene.
Irrelevant off-target sites are excluded
CRISPRroots considers all these possibilities so researchers can easily harness the power of RNA-seq in the evaluation of their CRISPR experiments. However, so far, many researchers have been limited in their attempts to identify off-target effects by not utilising RNA-seq, explains Giulia Corsi. She is the paper's first author and works as a bioinformatician in Gorodkin’s group. Corsi elaborates:
»Off-target prediction tools give you a list of about a thousand potential gRNA-binding sites in your reference genome and score them according to how well the gRNA complements a DNA sequence based on matches and mismatches or in terms of binding energies. Then it is up to the researchers to select the five or ten most likely off-target candidates and use DNA sequencing to check if these sites have been unintendedly edited. But these predicted off-target sites might turn out to be located in untranscribed regions of the genome that don't affect the cell even if they are edited. Alternatively, no editing might have occurred at the predicted off-target sites, so the genes are transcribed normally.«
“The CRISPRroots pipeline is designed to be as easy as possible to set up”Giulia Corsi
Since CRISPRroots adds a second and third layer of gene-editing assessment by incorporating two kinds of RNA-seq information - sequence variation and differential gene expression - many of the irrelevant potential off-target sites Giulia Corsi refers to can be excluded from the list. However, if CRISPRroots detects downregulation of a gene that is also predicted to be a possible off-target site based on sequence similarities, there is good reason to suspect that this is indeed the result of an off-target effect.
However, it is still possible that no off-target editing has taken place and that the observed downregulation is a secondary consequence of the on-target editing event. Therefore the researchers need to study in detail several predicted off-target sites. With CRISPRroots, however, the number of potential off-target sites will be smaller, and the chance of any of them being critical will be higher than with traditional prediction tools. Consequently, the researchers can save a lot of work and resources.
»The CRISPRroots pipeline is designed to be as easy as possible to set up. You only need to input the reference genome sequence and the sequences your gRNAs are designed to target - it takes five minutes - and the RNA-seq data is accepted unprocessed as it comes from the sequencing machine,« explains Corsi. The software tool then executes all processes and delivers the resulting Excel files in a number of hours that depends mostly on the number of replicates and the volume of sequencing data.
CRISPRroots finds what other tools miss
CRISPRroots was tested on data from five published studies that include seven RNA-seq datasets of CRISPR-edited and control samples. The studies included both CRISPR-Cas9 knock-in and knockout experiments, and CRISPRroots was used to identify potential off-target sites and assess on-target gene-editing activity.
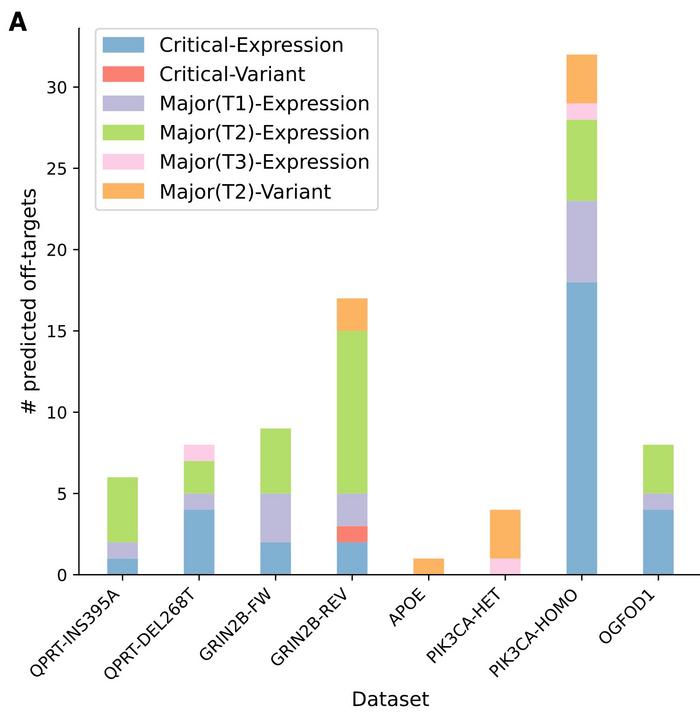
CRISPRroots identified potential critical off-target sites not picked up in the original studies in all seven datasets. The initial studies failed to detect them mainly because of limitations in the off-target prediction tools they used.
CRISPRroots incorporates an off-target prediction tool developed by Gorodkin and co-workers in 2018, CRISPRoff. Unlike most other tools, it considers elements like dG·rU and dT·rG wobble base pairs or non-canonical PAM sites such as NAG and NGA and generally leads to more accurate predictions. CRISPRroots reached more or less the same results as the original studies concerning on-target editing events. This was not surprising because edited sites are typically verified by sequencing.
Since the work on CRISPRroots has only just been published, the method has so far only been tested retrospectively on previous datasets, and other researchers have not yet used it to evaluate the results of ongoing CRISPR gene-editing experiments.
To conclude, the team behind CRISPRroots believes that other researchers will find their method useful since it can help them save time and resources in analysing their CRISPR gene-editing experiments.
Link to the original article in Nucleic Acids Research:
CRISPRroots: on- and off-target assessment of RNA-seq data in CRISPR–Cas9 edited cells
The CRISPRroot software tool is publicly available on https://rth.dk/resources/crispr/
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







