Diphtheria Toxin Selection Enables Site-Specific, Biallelic Gene Insertions

Gene editing is a valuable tool for creating disease models where two homogenous cell lines differ only in one gene of interest. Pharmaceutical companies use disease modelling in drug discovery, but creating the models can be difficult and time-consuming. It can easily take four to six weeks to edit and enrich the edited cell populations using traditional processes. Now, thanks to a novel method, it only takes a single week to do so at AstraZeneca.
Marcello Maresca, Senior Principal Scientist and Director at BioPharmaceuticals R&D, AstraZeneca, headed the development of the new CRISPR-based method. The method uses diphtheria toxin (DT) to select edited cells without introducing any exogenous genetic elements into the cells.
»We chose diphtheria toxin for its high specificity and potency. Ideally, we would like our selection reagents to eliminate all wild-type cells without affecting any edited cells,« explains Senior Research Scientist Songyuan Li. He is the first author of a recent paper in Nature Communications, where the new method is described.
Editing of Hbegf leads to DT resistance
DT exerts severe toxicity on cells when imported into cells via its receptor Hbegf which is located on the cell membrane. Li and his colleagues developed a gene-editing strategy that introduces mutations in this endogenous receptor to block its binding to DT. This effectively protects the edited cells from the entry of toxin molecules and, remarkably, does so without inserting any exogenous selection gene.
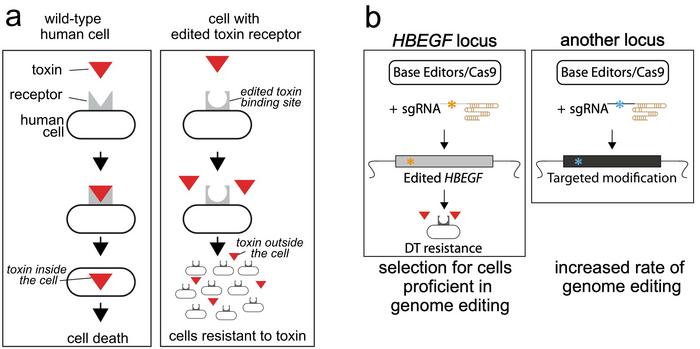
While human cells are sensitive to DT, this is not the case for murine cells due to slight genetic differences in the EFG-like domain of the Hbegf gene. So the research team initially wanted to see if they could introduce mouse-like mutations in the human gene that disrupt receptor/toxin binding and thus render the modified cells resistant to DT. To do this, they first designed 14 sgRNAs spanning the amino acids that differ between mouse and human at this locus.
Subsequently, they used base editing to introduce C-to-T or A-to-G mutations by co-expressing HEK293 cells with each of the sgRNAs and either of the two base editors CBE3 or ABE7.10. After exposure to DT, it turned out that cells transfected with CBE3/sgRNA10 and ABE7.10/sgRNA5 proliferated, while control cells and cells transfected with most of the other combinations died. This effectively demonstrated that editing of Hbegf could lead to DT resistance, and accordingly, the approach can be used to select for edited cells.
Clonal pools are easily obtained
The team reasoned that base editing of the Hbegf locus happens in the cells that are most proficient for editing. Accordingly, these edited cells might also be more permissive to DNA manipulation at another locus. In that case, DT selection could be used to co-select for the editing of another gene. The theory was tried out with co-transfection of either CBE or ABE base editors, or Cas9 with sgRNAs targeting Hbegf and several other genes, including Ctla4 and Pcsk9. The results showed that DT selection increased base conversion 4- to 13-fold and indel frequency 2- to 3-fold.
“Using this method, we can generate a clonal pool without clonal isolation”Songyuan Li
The next step was to modify the DT selection scheme for the enrichment of biallelic gene knock-ins using Cas9. Since DT resistance requires modification at both alleles, this can be achieved if the novel gene is inserted at the Hbegf locus.
»We designed sgRNAs and repair templates to introduce DT resistant mutations in the Hbegf intron together with any gene of interest. Only cells with biallelic resistant mutations introduced by DNA insertion will survive the treatment. Using this method, we can generate a clonal pool for homogenous gene expression without clonal isolation,« Li explains.
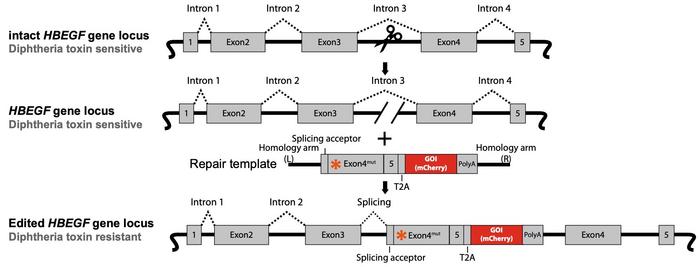
The template contained exon 4 of Hbegf containing the DT resistance mutation and exon 5 followed by the gene of interest, which initially was mCherry or GFP. Different versions of the template were designed with or without homology arms or flanking sgRNA cutting sites. This allowed for promoting integration by different modes of DNA repair.
»The results showed that our selection method can be applied to all types of DNA templates, including plasmid, dsDNA and ssDNA. Furthermore, it can be used for DNA insertions mediated by either homologous recombination (HR), non-homologous end-joining (NHEJ), or homology-mediated end joining (HMEJ). We see that templates promoting HR and HMEJ are the best choice for introducing biallelic DNA insertions at this locus in HEK293 cells,« Li explains. He points out that these templates generated almost complete insertion efficiency with GFP or mCherry fluorescence in nearly 100% of selected cells.
Xential enables co-selection of knock-out and knock-in editing
The researchers at AstraZeneca have given this system for precise genomic integration in the Hbegf locus the name Xential, an abbreviation for "recombination (X) at a locus conditionally essential for cell survival.”
One particular strength of Xential is that it provides biallelic gene insertion. This was demonstrated in an experiment where the repair template consisted of a puromycin resistance gene and a mCherry gene linked to the mutated Hbegf gene. After transfection, cells were selected using either puromycin or DT, and mCherry fluorescence was measured. In both cases, nearly 100% of selected cells were mCherry-positive, but fluorescence intensity was substantially higher in DT-selected cells. This is because knock-in at only one allele is sufficient for puromycin resistance, while knock-in at both alleles is required for resistance to DT.
Li and his colleagues also showed that Xential could be used to co-select knock-out or knock-in events at other genomic loci. Experiments performed with co-transfection of sgRNAs that targeted several genes showed that indel frequency increased knock-out 4- to 14-fold when non-selected cells were compared to DT-selected cells. Similar knock-in experiments resulted in a 4- to 6-fold increase in efficiency. Li summarises the results by saying:
»Xential potentially allows us to enrich cells with knock-ins or knock-outs at any genomic loci.«
“Xential potentially allows us to enrich cells with knock-in or knock-out at any genomic loci”Songyuan Li
While the initial experiments were performed with HEK293 cells, the team next investigated whether or not it could be used to enrich base editing in primary T cells and human iPSCs. These experiments showed that DT selection increased base editing efficiency in iPSCs 24- to 60-fold when targeting different genes, including Ctla4. In primary T cells, however, the increase was only 2-fold.
»This could be due to pre-existing DT cellular immunity in the primary T cells. To apply Xential in primary T cells, further engineering of DT or using synthetic immunotoxins will be needed,« says Li.
In vivo applications in humans require further modification
For clinical use, the edited Hbegf gene causing DT resistance must not perturb the gene’s normal cellular function. Hbegf is a membrane-anchored epidermal growth factor produced by monocytes and macrophages. It is involved in wound healing, cardiac hypertrophy, heart development and function, and cancer development.
“The mutations we introduced did not perturb Hbegf function, and we could not find any detectable effects on cell fitness or differentiation”Songyuan Li
»We checked the influence of resistant mutations on the function of Hbegf by analysing cell proliferation capacity and downstream signalling, without observing any significant difference between wild-type and mutated cells or proteins,« says Li. The team also checked the differentiation potential of mutated iPSCs and control iPSCs, observing no significant differences in their differentiation potentials to mesoderm, endoderm and ectoderm layers. Li concludes:
»The mutations we introduced did not perturb Hbegf function, and we could not find any detectable effects on cell fitness or differentiation.«
These promising results may allow Xential to be used in the clinic for ex vivo gene editing of human cells before transplanting them into the patient's body. But since DT is toxic to humans, DT selection cannot be used clinically in vivo without modifications to the method.
»We can see the need for and possibility of modifying the current method to be used in vivo. One option could be to replace DT with chimeric or synthetic immunotoxins or antibody-conjugated drugs with therapeutic value,« Li suggests.
Link to the original article in Nature Communications:
Universal toxin-based selection for precise genome engineering in human cells
Tags
ArticleNewsAdenovirus (AV)Quality ControlBase editorsCRISPR-CasCas9AstraZeneca
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







