DTMP-Prime Model Advances Prime Editing Predictions
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
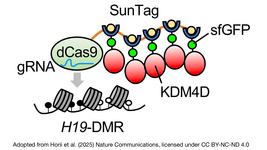
DTMP-Prime combines sophisticated feature selection, deep embedding, and multi-head attention layers to predict the efficiency of PE complexes by evaluating factors affecting pegRNA and target DNA sequence interactions. More than 43 features are used to enable the detailed analysis that reduces off-target effects and enhances editing outcomes, including pegRNA length, RNA folding structures, Cas9 activity, and nucleotide composition.
DTMP-Prime also uses DNABERT embeddings to capture nuanced relationships within DNA sequences, effectively identifying optimal pegRNA configurations for various PE systems, like PE2 and PE3. Performance comparisons with state-of-the-art PE prediction tools, such as DeepPE and Easy-Prime, demonstrate that DTMP-Prime achieves higher precision, with Pearson and Spearman correlation coefficients of 0.8 and 0.77, respectively, in test datasets.
This study was led by Leila Safari from the University of Zanjan, Iran, and Alireza Khanteymoori from the University of Freiburg. It was published today in Molecular Therapy: Nucleic Acids.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.