First Structure of the Tiny CRISPR-Cas12j3 Is Solved in Record Time
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Since CRISPR-Cas was first adapted for eukaryotic genome editing in 2013, the technology has advanced by leaps and bounds and the toolbox has grown to include additional Cas enzymes with distinct properties, and spin-off gene-editing techniques such as base editing and prime editing. Today, a number of CRISPR-based therapies are moving through clinical trials with promising results.
Despite these advances however, a number of challenges remain within the CRISPR medicine field, one of which is the challenge associated with the limited size capacity of the adeno-associated viral (AAV) vectors used to deliver gene-editing reagents to cells.
Last year’s discovery and initial characterisation of CasΦ (Cas12j) as a tiny Cas system that could edit mammalian genomes raised hopes that a solution to the AAV capacity challenge was within reach.
The latest work from Guillermo Montoya’s lab at the Novo Nordisk Foundation Center for Protein Research (CPR) at the University of Copenhagen in Denmark has brought that solution even closer. The group was behind a recent publication in Nature Communications describing in detail the structure of Cas12j3 (a member of the CasΦ (Cas12j) family) in complex with a target DNA after cleavage. This work provides the first insights into the molecular mechanism of action of a CasΦ family member, facilitating future redesign of this CRISPR system for therapeutic genome editing.
»As soon as we heard about CasΦ last year, we thought it was a very interesting molecular scissors. Its small size and the fact that it doesn’t need a tracer RNA makes it a very attractive nuclease, but we believed that to use it for targeted genome editing, we needed to understand exactly how it works. My team managed to do just this by solving the structure of Cas12j3 in record time, and just a few weeks later the group of Jennifer Doudna presented the structure of Cas12j2 (or CasΦ2), a closely related homologue«, said Guillermo Montoya, who is a Professor, Research Director and Group Leader at Novo Nordisk Foundation Centre, University of Copenhagen, Denmark.
Here, Guillermo Montoya is referring to a paper from Jennifer Doudna's lab published in Nature Structural & Molecular Biology on the 11th August 2021, just 3 weeks after Montoya's group published the work on Cas12j3.
Solving Cas12j structure – a simple yet complicated process
The team, which consisted of structural biologists, a microscopy specialist and molecular biologists, turned to cryo electron microscopy (cryo-EM) to solve the structure of Cas12j3, which is about half the size of the well-characterised Cas9 enzymes with a molecular weight in the range of 70-80 kDa.
Guillermo Montoya explains how the diverse team tackled the task of solving Cas12j3, which he describes as simple and complicated at the same time:
»We began by overexpressing the Cas12j3 protein in E. coli. We cloned it into an expression vector with a C-terminal 6-His tag that we could use for purification from bacterial cultures. We then used in vitro transcription to prepare the CRISPR RNA [crRNA] and used this to reconstitute the ribonucleoprotein (RNP) particle in vitro«.
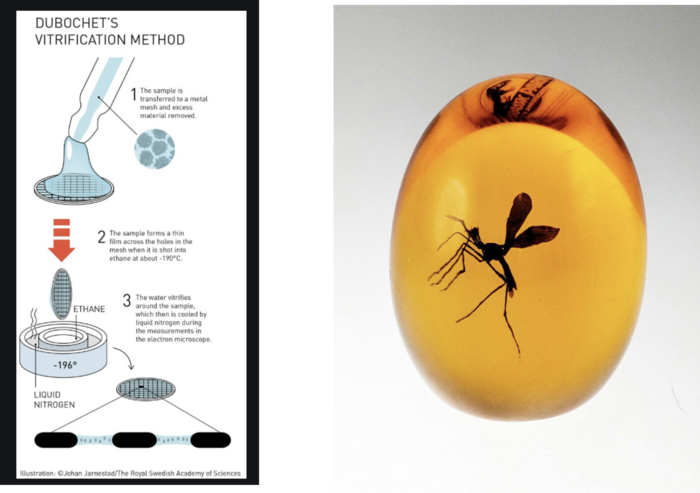
In the subsequent procedure, which is known as Dubochet’s vitrification method (Figure 1, left), the RNP particles are applied to a metal grid that is covered with carbon and which contains holes. The sample forms a thin film across the grid and excess material is removed. The remaining material on the grid is then flash-frozen to -190 °C in ethane, so that the RNP particles freeze and become embedded in a transparent phase, which is the aqueous buffer solution that remains vitreous.
The grid with the vitrified RNPs is exposed to the electron beam. The end result is many particles embedded in the vitreous ice, which visually resembles many insects that become trapped in a piece of developing amber (Figure 1, right).
Once the material is vitrified, it is inserted into an electron microscope. The protein particles in the vitrified grid will fall in different orientations, and each particle produces a trace on the image taken by the microscope. The attached computer is able to classify and group particles according to their orientations and similarities, and the in-built algorithm can generate high-resolution 2D images of the particles, which are then used to reconstruct a predictive 3D structure for the protein (Figure 2).
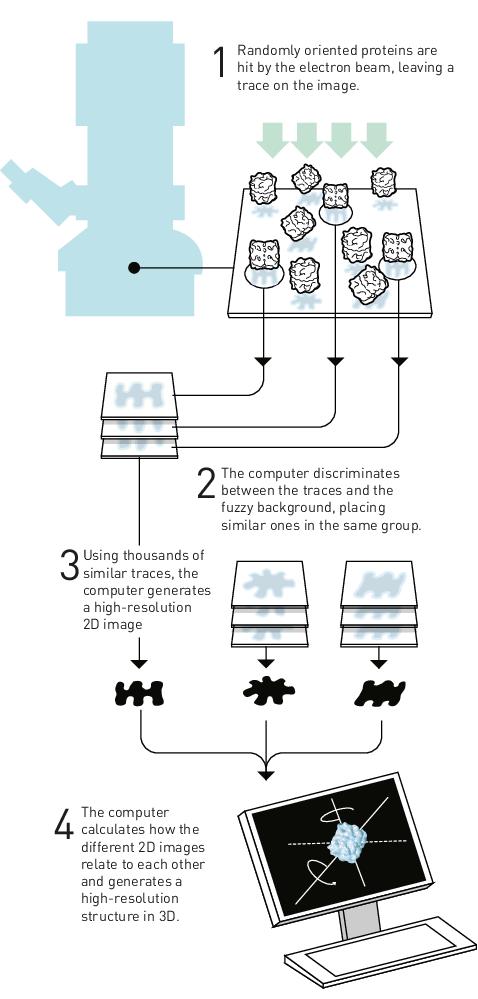
Cas12j3 structure reconstructed from high-resolution images
Montoya explains that the algorithms for cryo-EM are so good nowadays that if one is sure that they are applying the right sample to the grid, the structures that are returned are highly reliable.
One of the special properties of the cryo-EM approach is that you can take advantage of the sample conformational heterogeneity to solve several structures with one sample:
»When we prepare samples for cryo-EM the molecule undergoes conformational movements, and we can observe protein particles in different conformations in certain cases. This means that in some of the protein locations we don’t get a single structure, but we get several structures. This is very useful for getting an idea of the conformational changes of the molecule, in some cases while they carry out their function. We managed to do this when we solved the structure of Cas12a a few years back and we could do it here with Cas12j3. When we understand how the protein and its mutants looks in different conformations, we can begin to pinpoint regions that may be relevant for future redesign with respect to genome-editing applications«, explains Montoya.
Solving the structure opens the door to redesigning Cas12j3
Montoya’s team rationalised at the outset of the project that to realise the full potential of Cas12j3 as a compact genome editor, to be able to alter its specificity and the way it cleaves DNA, it was necessary to fully decipher its molecular mechanism of action including its protospacer adjacent motif (PAM) profile and how it interacts with its target. It turns out that they were right, as Montoya explains:
»Even though our main goal was to describe the mechanism of cleavage by Cas12j3, we also identified mutants with altered cleavage properties. Some of these variants will be very interesting for the genome-editing field. For instance, we identified one variant with an arginine mutated to a glutamic acid in the RuvC insertion region [the nuclease domain that initiates cleavage of the DNA strand], and this eliminated the possibility for single-stranded DNA cleavage while conserving the specific double-stranded DNA cleavage activity.«
Having solved the first structure of the CasΦ family, the team now has the information it needs to alter the cleavage properties of the tiny Cas12j3 nuclease. However, while making these nucleases more active is important to Montoya, the combination of the nuclease activity with improved precision is the key point:
»We hope that the recent structures of Cas12j3 and Cas12j2 will help to redesign this tool. We want these molecular scissors to cleave but not too much because this might induce off-target cleavage. I think what’s more important for us is the knowledge that we can eliminate certain properties such as non-specific ssDNA cleavage while retaining other more desirable properties such as specific dsDNA cleavage. Being able to do this with such a small nuclease is very attractive and this potential is something that we will continue to investigate.«
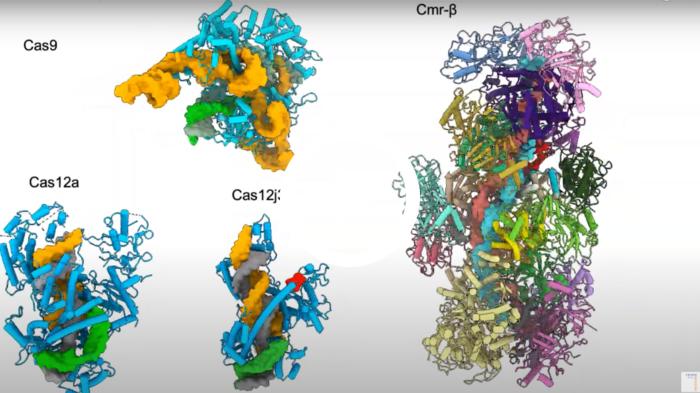
The video below illustrates the differences in size between Cas9, Cas12a and Cas12j3.
Video description: the structures of Cas9 (Jiang et al., Science 2016) Cas12a (Stella et al., Nature 2017), Cas12j3 (Carabias et al., Nature Comms 2021) and Cmr-beta, a CRISPR-Cas Type III effector complex, which is the largest effector complex characterised to date (Sofos et al., Molecular Cell 2020). The crRNA (orange) and T-strand DNA (grey) form the crRNA/target DNA hybrid. The hybrid size is similar for Cas12a and Cas12j3 while the Cas12j3 protein is half the size of Cas12a. Video courtesy of Guillermo Montoya.
Cas12j structure is a leap forward into new genome-editing applications
According to Guillermo Montoya, while some of the new Cas12j3 variants identified during the study have potential in applications that aim to inactivate a gene as well as introduce a new piece of DNA, their real potential may lie in applications that are not readily possible with the currently available Cas enzymes:
»Their reduced size frees up a lot of space in the AAV delivery vectors, so I think that we can start talking about applications that require the delivery of regulatory sequences as well as gene-editing cargo. This could open up for CRISPR-induced changes to epigenetic signalling and modulation of gene transcription rates, as well as simply inserting longer pieces of DNA than is currently possible.«
Room for many in the CRISPR toolbox
A few years ago, Guillermo Montoya’s lab solved the structure of Cas12a in multiple conformations, an RNA-guided endonuclease that recognises a different PAM than Cas9, has a smaller RNA than Cas9, and single catalytic sites for cutting each strand of a double helix.
Since its discovery, Cas12a has received much attention within CRISPR-based diagnostics, owing to its non-specific single-stranded DNA cleavage activity that it unleashed upon RNA-guided cleavage of a sequence-specific double-stranded DNA target. This property has been exploited by researchers to develop CRISPR-based detection assays where the non-specific cleavage of a labelled reporter molecule serves as a readout for the presence of the target sequence, e.g., viral RNA.
With structural insights into how Cas12a interacts with and cleaves DNA, the Montoya lab spunout this know-how to scientists at Twelve Bio to engineer superior variants for gene editing and diagnostic applications, including variants with non-specific ssDNA cleavage activity only, and variants with abolished ssDNA activity but retained dsDNA activity. The question is whether Cas12j3 can replace Cas12a? This is difficult to answer, according to Guillermo Montoya:
»This question comes up all the time. I’ve been working within genome editing for a long time now, and I worked with meganucleases and TALENs before I got into CRISPR-Cas. Both have been successfully used in different applications. When it comes to CRISPR, even though Cas9 is the most widely used nuclease and most data has been generated with Cas9, it is difficult for me to believe that the first Cas is going to be the one and only. I believe that all of these tools are going to be used in one way or another, as there are multiple possible applications. Cas12j family members will be another tool in the box.«
Link to original article in Nature Communications:
Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3.
Tags
ArticleInterviewNewsRibonucleoprotein (RNP)CasMINICRISPR-CasCasΦ
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.