Highlight: CRISPR Gene Editing Exposes Hidden Switch in Inflammatory Receptor
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
The story begins with a puzzle that has long challenged geneticists studying immune-mediated diseases. Genome-wide association studies repeatedly implicate regions near the TNFRSF1A gene in conditions such as ankylosing spondylitis, Crohn's disease, and multiple sclerosis. Yet most risk variants lie not within coding sequences, but in non-coding regions that influence how genes are switched on or off. Understanding what these regions do demands molecular tools of surgical precision.
“Understanding gene regulation and the genomic context in which GWAS variants lie could bring us closer to deconvoluting the genetic basis of common disease aetiology and uncover effective drug targets”Osgood et al.
Julie Osgood and colleagues at the University of Oxford turned to CRISPR-Cas9 to investigate a suspicious segment of DNA within the first intron of TNFRSF1A, which encodes the receptor for tumour necrosis factor alpha (TNFα) – one of immunology's most important signalling proteins (see Figure 1). Epigenomic profiling had revealed telltale marks of regulatory activity at this site: accessible chromatin, active histone modifications, and transcription-factor binding, all signatures of an enhancer that boosts gene expression.
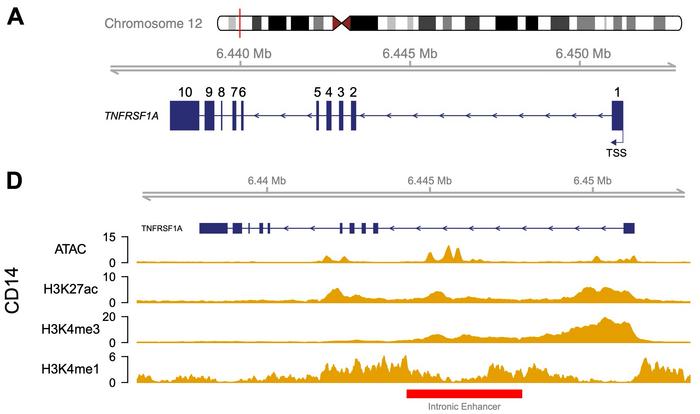
To test this directly, the team engineered a deletion of a 3.3 kb region encompassing the candidate enhancer in human induced pluripotent stem cells (iPSCs), then differentiated the edited lines into macrophages, immune cells that both produce and respond to TNFα. The authors state in their Scientific Reports paper:
"The aim of this work was to use CRISPR-Cas9 … for a functional interrogation of a candidate enhancer region in human induced pluripotent stem cells, followed by differentiation into macrophages, with functional genomic readouts including the assay for transposase-accessible chromatin using sequencing, chromatin immunoprecipitation with sequencing, and RNA-Seq used to assess the consequences of these edits."
The effects were striking yet confined. Chromatin accessibility and the histone mark H3K27ac disappeared precisely over the deleted region – the footprint of an active enhancer now erased. Genome-wide, no other regions changed significantly, confirming that CRISPR editing had not disrupted unrelated loci. RNA sequencing revealed a modest but statistically significant reduction in TNFRSF1A expression, while neighbouring genes within the same topologically associating domain were unaffected (see Figure 2).

Supporting experiments using a luciferase reporter assay showed that the deleted sequence could drive a roughly twenty-five-fold increase in reporter activity, providing independent evidence of enhancer function. Together, these results demonstrate that this short intronic region specifically enhances TNFRSF1A transcription in macrophages.
The deleted segment also overlaps predicted binding sites for transcription factors such as STAT3 and ETV7, which have previously been implicated in regulating TNFRSF1A. Variants within this enhancer have been linked to altered receptor expression and to genetic risk for autoimmune disorders, offering a plausible mechanistic bridge between non-coding variation and disease.
While the observed decrease in receptor mRNA was modest, the authors note that enhancer activity may vary with cellular state or stimulation, such as during inflammatory activation, and suggest that CRISPR interference or activation systems could refine these observations in future work. Even so, the chromatin and transcriptional data provide a consistent picture of local regulatory control.
The implications reach beyond basic gene regulation. TNF inhibitors are among the most prescribed biologic drugs for inflammatory diseases, yet responses vary widely between patients. The authors write:
"Understanding gene regulation and the genomic context in which GWAS variants lie could bring us closer to deconvoluting the genetic basis of common disease aetiology and uncover effective drug targets."
Understanding how TNFRSF1A expression is regulated in macrophages may inform why certain individuals respond better to TNF-blocking therapy.
The study demonstrates how CRISPR technology – combined with stem cell differentiation and multi-omic readouts – can advance functional genomics from correlation to causation. By precisely removing a non-coding element and tracking the molecular consequences, Osgood and colleagues have shown that a small intronic enhancer fine-tunes expression of one of the immune system's key receptors – a subtle control switch with far-reaching implications for inflammation and autoimmunity.
The study was led by Julie Osgood and Julian Knight from the University of Oxford. It was published in Scientific Reports on 7 October 2025.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleCMN HighlightsNewsCrohn's Inflammatory Bowel Disease, IBDMultiple SclerosisAutoimmune diseasesAutoimmune disease
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.