Improved RNA Editing Expands Gene Therapy Capabilities
RNA editing has emerged as a promising approach for therapeutic interventions for genetic diseases. Traditional systems primarily rely on adenosine deaminases acting on RNA (ADARs), which are enzymes that mediate the conversion of adenosine (A) to inosine (I) in double-stranded RNA (dsRNA). Now, American researchers have introduced a novel RNA-editing platform named DECOR (deaminase-enabled recoding of RNA), which acts on single-stranded RNA (ssRNA) and presents several advancements and differences compared to traditional ADAR-based systems (see Figure 1).
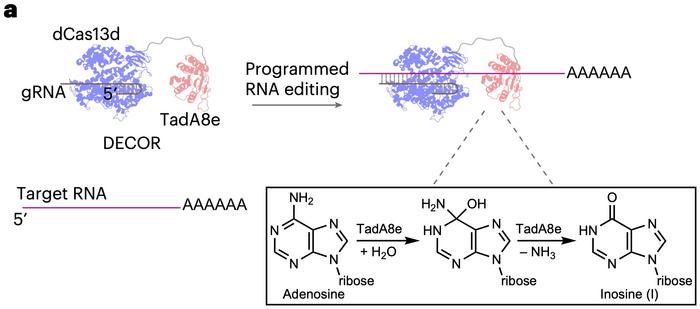
DECOR employs an evolved bacterial deaminase, TadA8e, fused to a catalytically inactive CRISPR-Cas13 (dCas13). This system allows for targeted A-to-I conversions within ssRNA regions specified by guide RNAs (gRNAs). The fusion of TadA8e with dCas13 ensures precise targeting and editing of RNA sequences without the need for dsRNA regions. The evolved TadA8e deaminase in DECOR has a relaxed sequence preference, enabling efficient editing across various RNA contexts while maintaining a low off-target profile.
DECOR’s effectiveness was demonstrated through a series of experiments across various human cell types, including HeLa (cervical cancer) and A549 (lung cancer) cell lines and human lung fibroblasts. In these experiments, DECOR targeted specific adenosines in RNA sequences such as EZH2-A1179, KRAS-A504, MALAT1-A2416, and METTL3-A950. The editing efficiency was quantified by measuring the conversion rates of A to I at the targeted sites.
In HeLa cells, DECOR delivered robust editing with conversion rates ranging from 18.1% to 26.9%, and in A549 cells, the rates ranged from 6.9% to 19.4%. In human lung fibroblasts, DECOR achieved a conversion rate of 3.0% for EZH2-A1179, significantly higher than the background editing level of 0.4% observed with a non-targeting guide RNA. These results confirmed that the observed RNA editing was specific and programmed by the guide RNAs.
DECOR’s performance was also benchmarked against existing CRISPR-based RNA-editing platforms, such as REPAIRv1 and its variants. DECOR consistently showed higher on-target editing efficiency and lower off-target effects, demonstrating its ability to function broadly in different human cell types, including primary cells. These abilities underscore its potential for precise and efficient RNA editing in therapeutic applications.
The clinical potential of DECOR was particularly highlighted by targeting the IRF6 gene, whose mutations are linked to Van der Woude syndrome, a genetic disorder. The IRF6 gene contains an upstream open reading frame (ORF) that negatively affects the expression of the primary open reading frame (pORF), leading to insufficient levels of the IRF6 protein.
Using DECOR, the researchers edited the uORF in the 5′ untranslated region (UTR) of the IRF6 mRNA. The editing removed the pathogenic uORF, thereby restoring the expression of the primary ORF. The results showed that DECOR-mediated RNA editing increased the expression of the primary ORF from 12.3% to 36.5% of the level seen in healthy transcripts.
The researchers also explored DECOR’s clinical versatility by applying it to other genes and conditions. For instance, DECOR targeted and edited sequences in the KRAS gene, which is often mutated in various cancers, demonstrating the system’s potential to address multiple genetic conditions. This adaptability highlights the broad applicability of DECOR in therapeutic settings, potentially offering new avenues for treating a wide range of genetic disorders.
This research was conducted by Hao Yan and Weixin Tang at the University of Chicago, and it was published in Nature Chemical Biology last Friday.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







