Massive CRISPR-Cas13 surveillance chip to fight COVID19 and future pandemics
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

A new study has just been rushed to publication in the high-impact journal Nature. It may be the key to stopping COVID19 and preventing future pandemics. The study describes a new system called CARMEN that allows fast and cheap testing for viruses on a massive scale with the impressive precision of CRISPR.
»We learned about the COVID19 outbreak while the paper was in review, and moved to develop a new assay for the SARS-CoV-2 virus quickly,« says senior author Paul Blainey, Broad Institute of MIT and Harvard.
He is accompanied by co-senior author Pardis Sabeti, investigator at Harvard University and Howard Hughes Medical Institute as well as co-first authors Cameron Myhrvold and Cheri Ackerman at an online press event.
»We submitted the paper prior to the outbreak and were excited to get it out. Now it is a very different feeling - a sense of incredible urgency to get it out to the world,« says Pardis Sabeti.
»Fundamentally, we hope it can help and impact this pandemic.«
The new CARMEN system is a powerful fusion of microfluidics, microarrays, and the newly developed CRISPR-Cas13 assay called SHERLOCK. In short, CARMEN runs thousands of nanolitre-scale SHERLOCK assays in parallel on a rubber chip not much larger than a smartphone.
It is a pathogen detection tool that allows surveillance of new infectious diseases to help fight pandemics now and in the future.
SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing)
CARMEN (Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic acids)
SHERLOCK detection miniaturised
The SHERLOCK assay was developed in the lab of Feng Zhang, which is also located at the Broad Institute. It builds on CRISPR-Cas13, which, like Cas9, can be programmed with CRISPR RNA (crRNA) to target RNA, e.g., from viruses such as SARS-COV-2. When Cas13 binds, it performs unspecific cleavage activity. This is not very useful for precision gene editing, but by adding a quenched fluorescent RNA reporter, the Zhang lab has turned it into a powerful detection tool. Here is a video for more information on SHERLOCK.
Now Paul Blainey and colleagues have found a way to implement the SHERLOCK assay chemistry in tiny nanolitre-scale microdroplets by adapting a drug discovery technology.
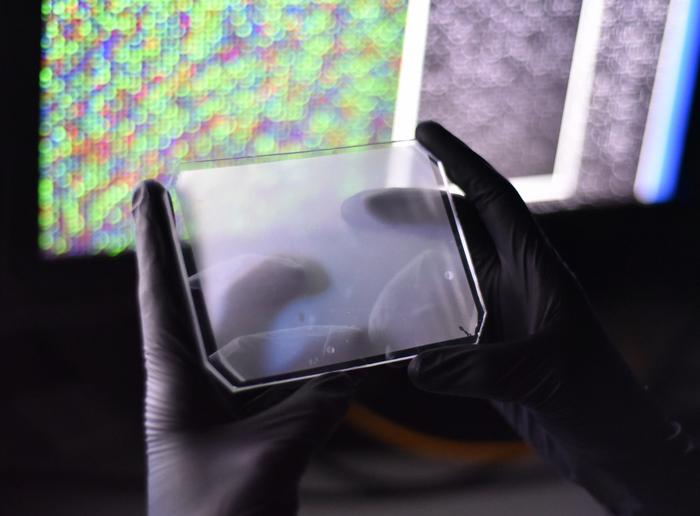
»We invented this platform a number of years ago for small molecule drug screening. Then we decided to repurpose the microfluidic platform to infectious disease diagnostics,« says Paul Blainey.
As the paper was in review, they learned about the new coronavirus outbreak, and it was relatively straightforward to incorporate a new test. They just had to design new crRNAs that target the SARS-CoV-2 virus, and within a few days they had a diagnostic test.
»We were excited when we got 80% precision on the first run and after some optimisation over 90%,« says first-author postdoc Cheri Ackerman.
CRISPR, nanofluids and microwells
The smartphone sized CARMEN chip runs about 4,000 tests and has more than 150,000 microwells, so each test is composed of many microwells. Each well accommodates exactly two microdroplets of emulsified samples containing either RNA or the detection reagents. To read the results, the researchers use a fluorescent microscope. All the individual droplets carry a colour code, and so from an image of the chip, the researchers can tell what sample and what reaction mix is in each well. To initiate the detection reactions, they apply an electric field to merge the droplet pairs in all the wells. When Cas13 finds a target, the fluorophore is released and detected as tiny green fluorescent spots.
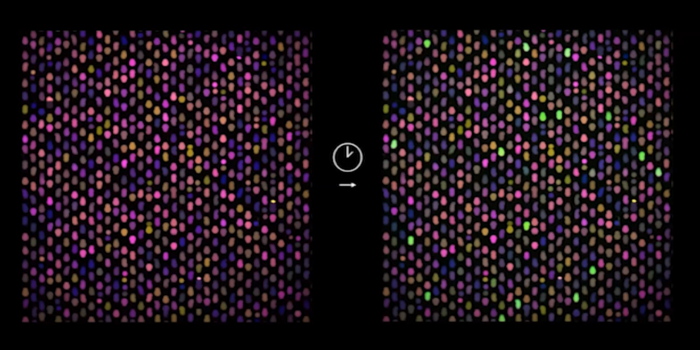
These are matched with the image before the reactions to identify which viral sequences have been detected in which samples. All of this can be done in a few hours.
Highly sensitive, robust and specific
And it works impressively well.
Sabeti and colleagues have previously demonstrated that SHERLOCK can detect viruses in patient samples down to attomolar concentrations.
Now, they show that the CARMEN system is just as sensitive and also that it can distinguish between different strains of viruses including Zika and subtypes of Influenza viruses, and it can even identify drug-resistant mutations in HIV.
In short, the technology is sensitive, specific, and statistically robust, matching the sensitivity of SHERLOCK and the standard PCR-based tests.
COVID19 surveillance
In the immediate future, the researchers want to deploy the CARMEN testing technology in the fight against COVID19. The researchers are currently discussing with the government and corporate partners how to do large-scale testing both in the U.S. and around the world.
»We see the role now as being in surveillance, either for clinical screening tests or for public health screening and surveillance testing,« says Paul Blainey.
The ability to test large numbers of patient samples can be a huge game-changer for stopping the current pandemic.
Testing is the window to understand the pandemic - how the virus is spreading, where it is and who is at risk, it may help to guide treatment, measure the effect of countermeasures, and so on.
In the longer term Paul Blainey imagines CARMEN could be deployed to test thousands of samples from selected populations, animal reservoirs, and patients presenting with symptoms. It can be part of a surveillance system for spotting new emerging infectious diseases and stopping outbreaks early.
»We hope to live in a world where we can identify any kind of infectious illness, so we can begin to figure out how to respond more quickly. So we can prevent pandemics and not just react to them,« says Pardis Sabeti.
In the doctor's office
The CARMEN system could also have a significant impact on our public health system in general.
»Imagine a world in which you go to the doctor, and you actually find out which virus is making you sick,« says Cheri Ackerman.
»So not a situation where the doctor tells you 'oh you have a viral infection, just go home and sleep it off'. You actually find out what virus you have and what medicine to take.«
She envisions that it could change public health if applied widely to every person having an infection.
»Imagine how that would impact epidemiology. Now, instead of wondering what virus is circulating in a community, we get statistics - real good measurements of what is there, and how fast is it spreading,« says Ackerman.
»I think that it is critical to have that kind of information. It continues to motivate us, and we are working hard to move this forward into the clinic, and impact patients and public health.«
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







