Minimal Cas9 ancestor enables durable epigenetic control
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
The study addresses longstanding limitations in genome editors by enhancing the activity and specificity of the IscB enzyme, a minimal ancestor of Cas9, while maintaining a compact size suitable for adeno-associated virus (AAV) packaging. The research team from the Broad Institute of MIT and Harvard screened diverse IscB orthologs and used structure-guided domain insertions, rational mutagenesis, and loop recombination to increase guide RNA recognition and reduce off-target effects.
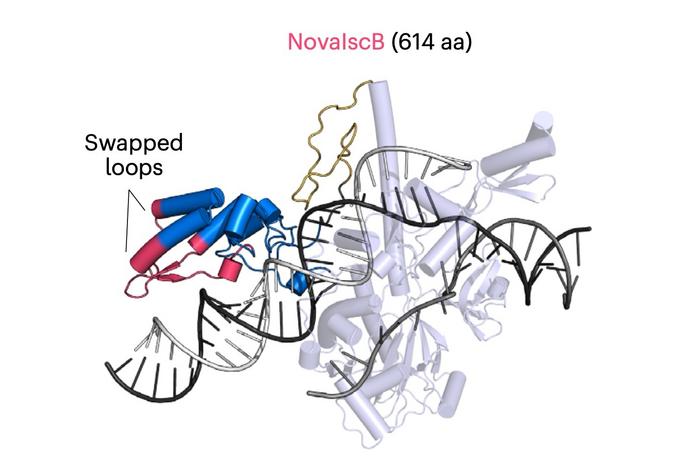
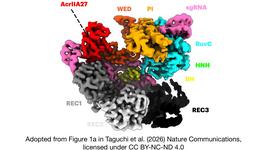
NovaIscB achieved a ~100-fold activity improvement over the original OgeuIscB, with a shift in effective guide length from ~13 to ~16–20 nucleotides, enhancing targeting specificity. Structural analysis via cryo-EM revealed extended duplex recognition and unique catalytic features distinct from Cas9, such as a tripartite histidine cluster coordinating a single Mg²⁺ ion in the HNH domain.
The NovaIscB system was fused with a DNA methyltransferase and KRAB domain to generate OMEGAoff, a transcriptional repressor capable of stable gene silencing. In both cultured cells and mouse models, OMEGAoff delivered durable repression of genes, such as Pcsk9, with reductions in protein levels and serum cholesterol. A truncated ωRNA scaffold further facilitated AAV packaging and increased expression efficiency, enabling robust in vivo delivery.
Feng Zhang led this work at the Broad Institute of MIT and Harvard, and it was published in Nature Biotechnology on 7 May 2025.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.