New platform enables efficient and safe CRISPR tools for gene-editing therapies

CRISPR-Cas has recently become an almost universal genome editing technique for molecular biologists, and with its growing popularity, scientists have found multiple ways to exploit it. While the first purpose was to cut and edit a gene, CRISPR is now used extensively to edit genes without cutting, process RNA rather than DNA, promote transcription, alter epigenetic markers, unleash fluorescent signals etc. The new possibilities have caused the CRISPR toolbox to expand from the original Cas9 isolated from Streptococcus pyogenes to several new Cas variants found in other bacteria as well as engineered versions of these enzymes.
So whenever scientists prepare for a new experiment, they need to decide on which Cas variant to use and that can be tricky, since all the variants have different enzyme kinetics that will influence the experiment for better or worse. To simplify the choice, a group of scientists from the Ilya Finkelstein Lab at the University of Texas at Austin, US has established a platform that allows for massive parallel kinetic profiling of CRISPR nucleases. The platform is called NucleaSeq and is presented today in Nature Biotechnology. CRISPR Medicine News had the chance to interview the two leading authors, Stephen Jones and John Hawkins.
“The name of the game in CRISPR-based medicine is having safe, controllable results. This work is a significant step along that path, in both the short term and long term”John Hawkins,
»The name of the game in CRISPR-based medicine is having safe, controllable results. This work is a significant step along that path, in both the short term and long term,« says Hawkins and continues: »In the short term, this method will help researchers quickly and efficiently choose the right CRISPR nucleases (among dozens) and DNA targets (among thousands) for a given application, producing better results, and doing so faster and cheaper than previously possible.«
NucleaSeq reveals standardised benchmarks of Cas nucleases
Most of the information we now have on cleavage and binding specificity of Cas variants rely on experiments with specific targets in genomic DNA. But when you do the math, it turns out that the genome is not nearly big enough to look at all possible off-targets. Cas nucleases will ignore sequences completely if they differ from the target by just a few bp, so in the genome there are typically only a couple dozen sites that are potential mismatches. These will only cover a fraction of all the possible mismatches that you need to test your Cas variant against. To solve this problem, the NucleaSeq platform is testing the nucleases against around 10,000 synthetic DNA sequences containing predicted mismatches, insertions and deletions relative to the guide RNA and thus have the ability to provide the highest information content. The massive number of nuclease reactions are carried out in solution and cleaved products are captured over time and subjected to deep sequencing.
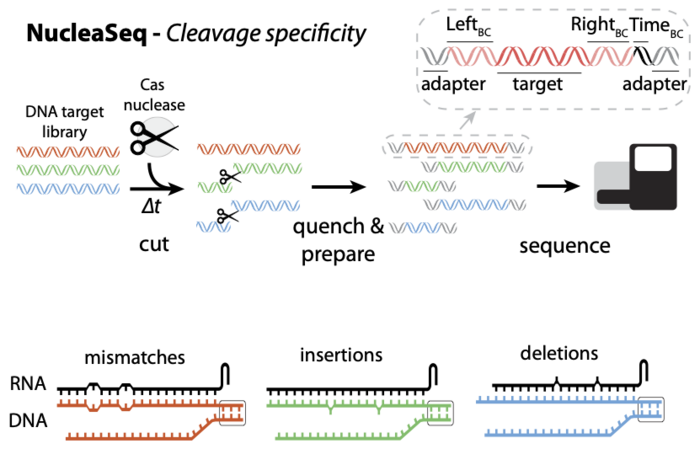
NucleaSeq reveals several standardised benchmarks of the Cas nucleases tested, and Jones says: »This is the only way we can reliably compare them.« As an example, he points out that while in vivo-experiments usually find that Cas12a appears to edit fewer off-targets than Cas9, NucleaSeq found the two enzymes to have similar cleavage specificities.
»We think this is because Cas12a is a bit slower overall, so cellular enzymes may be able to knock it off of off-targets before it cleaves them. Those types of factors can depend heavily on the cell, the target and even the exposure time,« he explains.
CRISPR users must evaluate cleavage specificity vs binding specificity
The two scientists believe the NucleaSeq platform will enable other researchers to choose the best Cas variant for their experiments. Ideally, researchers want a CRISPR protein that will find its target site directly, without lingering anywhere else in the genome. Once there, the preferred behaviour depends on the application. For correcting genetic diseases, you would want precise cuts that cleanly facilitate a change to the right genetic sequence. For research, often what you want is to mess things up at a specific location in order to see what happens. And for binding applications, you want to be able to tune how much time the Cas spends bound to the target site versus how much time it gives access to other proteins.
»There are many applications for CRISPR technology, but they basically fall into two camps: whether you want to modify the DNA or you want to interact with the DNA in some other way,« Hawkins explains.
»Applications for modifying the genome - to remove the cause of a genetic disease, for example - typically require DNA cleavage to cut out the original sequence. Applications to interact with the genome - say, to promote expression of a gene of interest or to get in the way of some other DNA binding protein - typically only require DNA binding,« the bioinformatician adds.
Jones, a molecular biologist, elaborates: »Cleavage specificity is important for editing: It describes where and how you can expect the nuclease to cut DNA - i.e. at your target and related off-targets, and whether you will get blunt or staggered DNA ends. Binding specificity, on the other hand, is important for localisation: It describes if and where you can expect the nuclease to bind DNA - there are many more places that they bind than where they cleave.«
Tailor-made CRISPR nucleases are expected in the future
The two scientists believe their new, massively parallel screening platform has great potential for the future. »In the long term, NucleaSeq will probably provide in-depth performance results for each CRISPR protein in a way that points to specific places in the protein where modifications could improve performance. We believe this will empower the design of a whole new generation of engineered CRISPR proteins with better performance than is possible today,« says Hawkins.
And Jones adds: »We're working on ways to make NucleaSeq even quicker and easier to use, so stay tuned!«
Link to the original article in Nature Biotechnology: Massively parallel kinetic profiling of natural and engineered CRISPR nucleases.
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University