New Tool Enables Allele-Specific CRISPR Editing
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
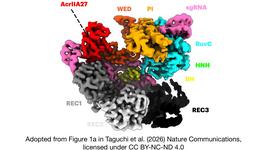
The study addresses the challenge of distinguishing single-nucleotide variations (SNVs) in a pathogenic genomic context for dominantly inherited genetic diseases. AlPaCas automates the identification of SNV-derived PAMs, which are crucial for precise targeting in CRISPR editing. By inputting gene or SNV data, users can receive a list of potential PAMs specific to the mutant allele.
The web server then cross-references these with a comprehensive database of PAM patterns associated with various Cas enzymes. This enables the selection of the most appropriate enzyme for allele-specific editing, ensuring that the wild-type allele remains unaffected. The identification process is complemented by structural analysis, offering insights into the interaction between the Cas enzyme and the SNV-derived PAM.
The innovation lies in the server’s ability to propose mutational engineering of Cas enzymes to enhance their selectivity towards the mutant allele. This involves modifying amino acids in the PAM-interacting residues of the Cas enzyme, guided by evolutionary conservation and binding energy calculations.
Such modifications increase the enzyme’s affinity for the mutant allele’s PAM while reducing off-target effects on the wild-type allele. The AlPaCas tool also includes a user-friendly interface, making it accessible to researchers with varying levels of expertise. This computational resource represents a significant advancement in precision gene editing, potentially transforming therapeutic approaches for genetic disorders.
AlPaCas is developed by researchers from Sapienza University of Rome and the University of Modena. The study was published this weekend in Nucleic Acids Research.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.