Next-Generation CRISPR Screening Libraries Without PCR Bias and Cloning Hassle – Interview with Vivlion
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

(Interview is condensed and edited for clarity)
- Hi Manuel and Martin. It’s great to speak with you both today. I’d like to start with the basics. What exactly is meant by a CRISPR screen?
Martin: A CRISPR screen is a functional genomics experiment that can reveal clues about the functions of genes in a specific context. Typically, a library of plasmids, each containing one or more guide RNAs (gRNAs) targeting the investigated genes, is delivered to cells, along with a nuclease such as Cas9. Depending on the nuclease supplied, this results in a population of cells where one or more genes are edited to be knocked out, inhibited, activated, or changed in their amino acid composition. Among other uses, CRISPR screens are widely performed to investigate whether certain genes are involved in disease progression or in mediating resistance or susceptibility to certain drugs.
Manuel: The human genome encompasses about 20,000 protein-coding genes, and it’s often not clear what a given gene, or a combination of genes, does in a particular context, for instance, the contribution of a particular set of genes to a disease phenotype. Depending on how they are designed and executed, CRISPR screens can allow us, in a non-biased way, to look at and understand the function of single genes or gene combinations in a cell population or within individual cells in specific contexts or conditions.
Two main approaches to CRISPR screening
- CRISPR screens seem to be popping up in the literature on a weekly if not daily basis, and I’ve noticed much variety in the strategies and workflows being used. Can you take us through the most common approaches?
Manuel: CRISPR screening has been available for about 10 years now and it has been applied in many ways, but in essence there are two conventional approaches: CRISPR dropout screens and CRISPR enrichment screens. These forward screening approaches are independent of the biological question being asked and are widely used to identify genes that cause, for example, drug susceptibility or drug resistance in cancer cells.
In dropout screens (or negative-selection screens), cells containing gRNAs that target essential genes will be depleted from the pool of cells, while editing of genes that mediate drug resistance will increase the cells’ susceptibility to that drug. Conversely, positive-selection or enrichment screens reveal selective advantages such as drug or toxin resistance when specific genes are edited.
“You’re doing an experiment without any prior knowledge, and you let biology reveal itself without any preconception about the potential outcome.”Manuel Kaulich
Within the research field, many groups choose to include a fluorescent reporter for a specific biochemical process; this allows them to screen for factors that either activate that reporter (positive selection) or repress that reporter (negative selection).
So, in a nutshell, depleted and enriched cell populations are the essence of CRISPR screening. The experimental question being asked will determine which of those populations is most interesting. In practice, screens vary according to how the screening library is prepared, the number of gRNAs used, whether single or multiple transcripts are overexpressed (CRISPR activation) or suppressed (CRISPR inhibition), and depending on the gene-editing modality used, but these subtle variations of the technology do not change the core concept of cell populations being enriched or depleted.
The problems with standard CRISPR libraries – lack of uniformity and bias
- My understanding is that while CRISPR screens are powerful, they can also be very laborious and bias is a concern. Can you walk us through the main challenges with CRISPR screening?
Martin: Challenges arise across three main areas that must be addressed at the outset of any screening experiment. These are experimental design, library preparation, and identifying robust controls.
- Let’s talk about the challenges surrounding library design. What is crucial here and where are the pitfalls?
Manuel: There are three main issues here: completeness, gRNA distribution and size of the experiment. The main questions during the planning phase are: are you really investigating all of the genes that you wish to investigate? Are the gRNAs evenly distributed within the screening library? Is the size of the experiment manageable?
Let’s take a genome-wide screen in human cells as an example. This involves 20,000 genes and the overall experiment size will depend on how many gRNAs are used to target each gene. Ideally, each guide should be represented uniformly in the library, but this is not guaranteed with the way standard CRISPR libraries are prepared.
For standard library preparation, commercial oligonucleotide synthesis technologies are used to generate gRNAs pools. These are uniformly distributed when they come off the synthesiser, but they are then amplified using PCR and cloned into viral vectors for screening. Because the PCR step introduces bias, the obtained library can have undesired properties such as an incomplete and non-uniform gRNA representation. When it comes to screening and sequencing the gRNAs present in the investigated cells at the end of a screen, non-uniform libraries can require up to 1,000-fold coverage in order to detect low-abundance guides. A uniform screening library allows for a much smaller experiment without the risk of missing potential hits while maintaining or even improving data quality.
Vivlion’s CRISPR libraries are uniquely produced without any PCR step. This allows users to maintain the distribution of the original gRNA pool and transfer it directly into the experiment. Because our libraries are uniformly distributed regardless of library size, experimental size and the amount of cell culture work needed is significantly reduced (up to 10x). This allows researchers to perform CRISPR screens at scale, with sparse cell material (highly beneficial for in vivo experiments), and results in cost savings due to the lower cell maintenance and sequencing coverage needed.
Vivlion’s PRCISR™ CRISPR libraries allow complete, uniform and compact experiments
- How are Vivlion’s CRISPR libraries generated?
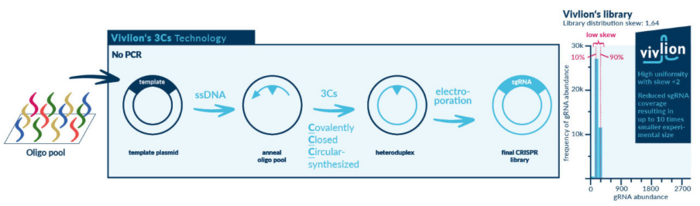
Martin: We generate libraries using our Covalently-Closed-Circular-synthesized (3Cs) technology, which enables a PCR-free and cloning-free workflow.
We start with a plasmid that contains an F1 origin; this is a phage-derived origin of replication that is present in most plasmids. Using phages and a special strain of bacteria, we generate a single-stranded version of the plasmid that randomly contains uracil instead of thymine, and design a library of gRNA-encoding oligos that can site-specifically anneal to a template sequence in the single-stranded DNA that serves as a gRNA placeholder. This in vitro reaction takes about 10-15 mins.
We then add a polymerase that extends the annealed oligonucleotides along the template plasmid, and a subsequent ligation step leads to the formation of a heteroduplex of the new and old strands. The heteroduplex is transformed into bacteria that are base-excision repair proficient. Since the template strand contains uracil at random positions, it is recognised and degraded, which leads to the preferential amplification of the gRNA-containing strand. This results in a plasmid DNA library that contains the desired gRNAs. Since the degradation of the template strand in bacteria is not perfect, we use a molecular biological trick to remove it completely: The template strand that initially served as a placeholder for the gRNAs contains a long restriction site, which we can use to easily remove any remaining template plasmids from our library by restriction digestion. The result is a PRCISR™ CRISPR gRNA library.
One of the major benefits of our approach is that library diversity is governed solely by the number of gRNA-encoding oligos. The distribution of oligos remains unchanged throughout library generation, so that the uniformity in a synthesised oligo pool is preserved in the final library.
Curated CRISPR libraries for human and mouse whole-genome screens
- How does Vivlion choose which gRNAs go into the libraries?
Manuel: CRISPR has been accessible for roughly 12 years, and there are probably few human cell lines left that have not been subjected to a CRISPR screen. This means there is a vast amount of publicly available CRISPR screening data. We have used a combination of these data, gold-standard machine learning algorithms, and validation experiments to identify the best gRNAs, i.e., the guides that exhibit target specificity and result in the most robust and reproducible phenotypes across cell lines and experimental contexts.
Vivlion’s single PRCISR™ CRISPR libraries generally contain four gRNAs for each gene in the human or mouse genome, with minimal variance within each target-specific gRNA set. For combinatorial libraries, the number of gRNAs per gene can vary depending on the experimental scale. However, the number and design of our gRNAs makes it easier to identify genes that are truly responsible for the observed phenotype within a screen, thereby adding confidence to the data.
- What level of documentation is supplied with the PRCISR™ libraries?
Each PRCISR™ CRISPR library is subjected to rigorous QC through molecular biology techniques and next-generation sequencing (NGS). Plus, we provide analytical QC reports and NGS read-counts with each library so that the user can directly start the experiment without the need to repeat the QC step.
Experimental design and proper controls are crucial to screening success
- Besides the library challenges we have discussed, you mentioned earlier that challenges also arise within experimental design and identifying the right controls. What are the crucial considerations for experimental design?
Martin: Within the experimental design phase, the hypothesis or biological question being asked should be properly defined so that the experiment can be set up in the best possible way to answer it. For us, project success is the most important measure. That also means that we point clients to alternative solutions if we think they are superior to our services. It is also crucial to ensure that the data eventually coming out of the screen can be analysed adequately and have enough statistical power to be meaningful. For us, this means including data scientists early on in the library and screen design phase and discussing a proper control concept for the screening experiment, and, if required, also for the library.
Size and scale of the experiment should also be considered. When it is not possible to screen all the desired genes or gene combinations in a single experiment, one must downscale such that the most interesting gene combinations or sets of genes are prioritised.
- Is there a general rule of thumb when it comes to identifying controls for CRISPR screens?
Martin: Two types of controls must be included in every CRISPR screen. Firstly, one must include positive- and negative phenotype controls. These are used to confirm that the gene-editing reagents are being delivered correctly to the cells and that the editing conditions are optimally set up, and to establish a baseline of the measured phenotype at the end of the screen, respectively.
Secondly, positive editing controls are usually validated gRNAs that have demonstrated high editing efficiency across multiple cell types without inducing a phenotype. Non-targeting guides that have low sequence similarity to the genome being targeted typically serve as negative editing controls, but these should allow loading with Cas9 (or whatever nuclease is being used); this way, the negative control is mechanistically similar to the targeting and positive-control gRNAs.
Experimental controls help the researcher to minimise the influence of variables beyond the measured phenotypes. For instance, a dropout screen to detect drug sensitising genes should include untreated cells that are screened in parallel with the same library, such that conclusions can be made about the impact of gene edits and ultimately about phenotypes in the presence of the drug.
To sum up, proper preparation can result in good quality data, and errors or omissions during the design phase will likely lead to inconclusive data.
Standardisation is emerging within the CRISPR screening community
- Standard terminology, methods and tools are emerging in the genome-editing field, which is spearheaded by the NIST Genome Editing Consortium. With so many considerations and steps involved, is there a push for standardisation or benchmarking within the CRISPR screening community?
Manuel: The short answer is yes. There are four important parameters to look at when judging screen quality. These are: the number of genes being targeted, library quality, size of the experiment, and the computational methods used to analyse the data.
Pioneering research groups have performed systematic CRISPR screens in many different cell lines. Based on these and similar data, they extracted a core set of genes that result in conserved phenotypes when edited, regardless of cell type or experimental context. Among these are so-called core essential genes, and one of the QC steps in every CRISPR screening experiment should be to confirm that these essential, but also non-essential, genes perform as expected, i.e., are they really dropping out, and if so to what extent? This is a key QC step in judging the quality of a CRISPR screen.
RNA targeting screens and custom CRISPR libraries
- It sounds like the field is heading in the right direction. Before we finish up, what can we expect to see from Vivlion in the near future?
Manuel: There are a couple of developments we are fairly excited about. I’ll focus on our screening services, and I will let Martin conclude with our efforts regarding innovative reagents.
So, Cas 9 screens are great for identifying essential genes, with knockout leading to dropouts, but targeting transcripts is a really good approach to identifying essential genes that may also be drug sensitisers, since these would be ideal drug targets. So, if we expose cells to a drug at a concentration that causes a 50 % reduction in viability and perform an RNA-targeted screen at the same time, we expect to see more severe phenotypes in cells where a drug sensitiser is edited. We are very excited about what these types of projects can reveal!
We also actively explore combinatorial gene effects. Beyond identifying paralogues, these approaches provide the much needed resolution on gene synergy and can even be used to predict functionally active multi-protein complexes. Coupled to, let’s say, fluorescent reporters for specific biochemical processes, these efforts have the potential to reveal and explain highly complex biological processes and may even transform our understanding of certain biological concepts.
Martin: On the reagent side, right now, we are about to release up-to-date versions of mouse and human genome-scale libraries that are ready-to-use, and will add additional species in the very near future. In addition, we regularly supply custom libraries in various different formats and backbones, for instance, to clients who wish to use their own custom set of genes or gRNA sequences.
We also offer the full-service portfolio, meaning that we provide PRCISR™ CRISPR libraries and also perform screens including the analysis for clients. At the moment, we offer enrichment and depletion screens as Manuel said earlier, in formats of reporter screens, FACS-based enrichment screens, drug sensitiser screens, and many others.
We also help clients to select suitable algorithms for gRNA design and regularly collaborate with our clients to generate libraries for other model organisms beyond mouse and human. It may be trickier to design libraries for infrequently used model organisms, depending on how much data is already available, but we enjoy challenges and welcome informal queries from anyone who might be interested in custom libraries or screens.
Our latest product goes beyond CRISPR libraries and screens and solves the bottleneck of efficient homology-directed-repair. Here, we leverage our 3Cs expertise in generating circular-single-stranded (css) DNA to provide cssDNA to clients in sizes of up to 15-20kb. cssDNA supports highly efficient single- and multi-locus recombination without the activation of the innate immune response, which makes this type of reagent particularly suited when working with primary cells and for potential therapeutic applications. We plan to launch cssDNA for the broad market in Q2 2024 and are open for early requests.
- Thank you so much for taking the time to speak with me today. It sounds like you have some exciting times ahead!
Learn more and get in touch with Vivlion
To learn more about Vivlion’s 3Cs technology and CRISPR screening services, check out the website here, or get in touch with the team for an informal chat by sending an email to: info@vivlion.com.
Watch and hear Martin Wegner present Vivlion's PRCSR CRISPR technology in our previous CMN webinar entitled: CRISPR-based Genetic Screens Enable the Discovery of Oncology Drug Targets, Predictive Biomarkers, and Mechanisms of Resistance. Watch the on-demand webinar here.
Relevant publications
Key articles about 3Cs technology:
- Diehl V, Wegner M, Grumati P, et al. Minimized combinatorial CRISPR screens identify genetic interactions in autophagy. Nucleic Acids Res. 2021 Jun 4;49(10):5684-5704.
- Wegner M, Diehl V, Bittl V, et al. Circular synthesized CRISPR/Cas gRNAs for functional interrogations in the coding and noncoding genome. Elife. 2019 Mar 6;8: e42549.
Applications of 3Cs technology:
- Hsia O, Hinterndorfer M, Cowan AD, et al. Targeted protein degradation via intramolecular bivalent glues. Nature. 2024 Mar;627(8002):204-211.
- Wegner M, Kaulich M. ReCo: automated NGS read-counting of single and combinatorial CRISPR gRNAs. Bioinformatics. 2023 Aug 1;39(8): btad448.
- Cetin R, Wegner M, Luwisch L, et al. Optimized metrics for orthogonal combinatorial CRISPR screens. Sci Rep. 2023 May 6;13(1):7405.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.