Prime Editing: A Double-Strand Break Free Solution for Safe and Efficient Gene Editing
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Gene editing entails the precise manipulation of the information contained within the DNA sequence via molecular scissors, best known as programmable nucleases (PNs), e.g., Meganucleases, ZFNs, TALENs and Cas endonucleases.
In recent years, an increasing number of human clinical trials involving gene editing has started. Some of them already show promising results, as proved by the current sickle cell disease (SCD) and beta thalassemia (BT) trials.
Despite these achievements, researchers have not stopped there. They continue to look for novel strategies to perform gene editing, particularly those that do not require the introduction of double-stranded breaks (DSBs) within the genomic DNA.
Double stranded-break concerns
All the genome-editing modalities involving PNs, including CRISPR-based approaches, involve the generation of a DSB at the target site. Once the DSB occurs, the researchers take advantage of cellular DNA repair mechanisms to abrogate gene expression by gene knockout (KO) or to introduce a specific modification, such as correcting a point mutation back to its healthy form.
However, a DSB is one of the most troublesome genetic lesions a cell can experience, and if not properly repaired the consequences may be extreme, for example the recently described gross-chromosomal aberrations following CRISPR-Cas9 genome editing or even cell death.
Concerns linking DSB response and CRISPR have risen in particular after two publications in 2018 highlighted how CRISPR editing may select for cells with aberrant p53 expression. P53 is one of the most known and studied oncogenes. Although no major follow-ups came after these two publications, it was clear that a DSB-free approach was highly desirable to make CRISPR technology safer.
PRIME Editing: freeing CRISPR from double-stranded breaks
The first response to this need was base editors which combined a Cas9 able to cut only one of the two DNA strands – thus dubbed as Cas9 nickase (nCas9) – with specific enzymes able to seamlessly generate specific DNA base conversions.
Although very efficient, base editors are limited to a couple of the possible point mutations that can be introduced. Furthermore, base editors cannot create deletions or insertions, thus further limiting their application.
This is where prime editing enters the arena.
Prime editing is based on an elegant strategy that combines an nCas9 with a reverse transcriptase (RT). RT is an enzyme used to generate complementary DNA (cDNA) from an RNA template, in a process known as reverse transcription.
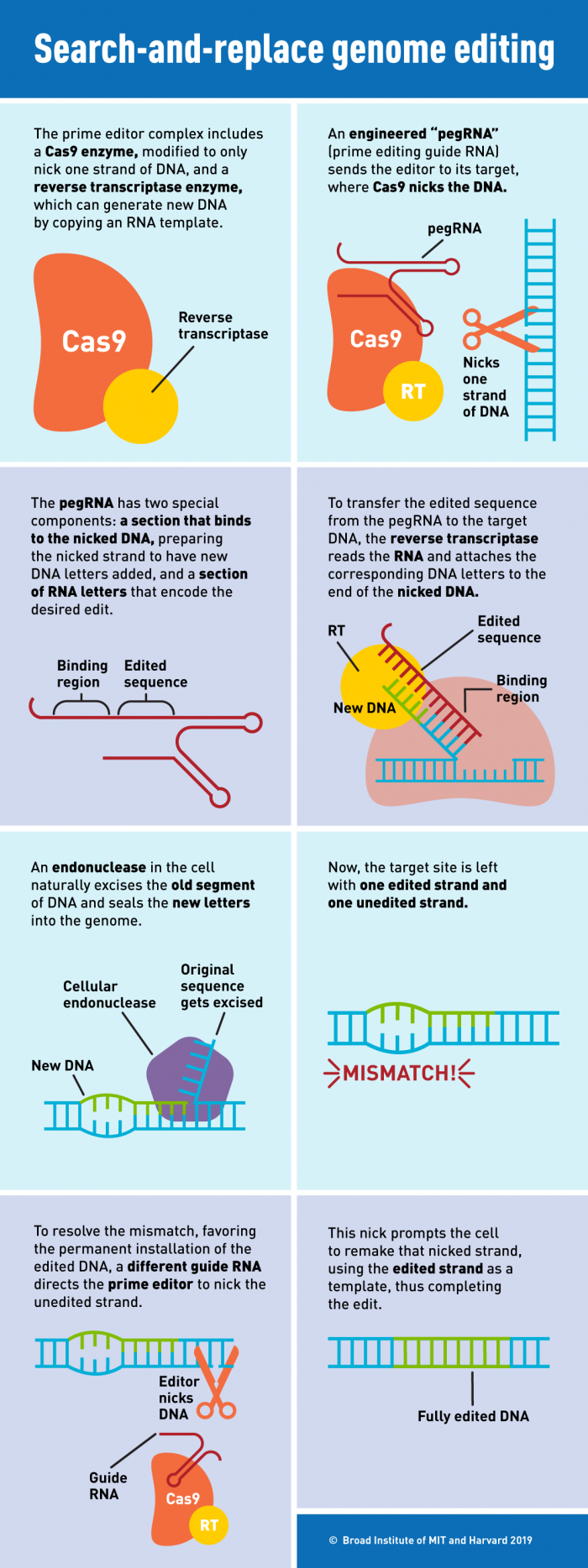
Prime editing makes use of a special guide RNA named pegRNA. The pegRNA acts both as the GPS that directs the nCas9 toward the genomic target and as the template itself. Besides possessing the sequence complementary to its target, a pegRNA also includes an RNA donor sequence. Once the nCas9 cuts a single strand of the DNA, the “donor component“ can be used as a template by the RT fused to the nCas9, which will transcribe a cDNA strand based on the RNA template included in the pegRNA. This newly synthesised cDNA will contain the desired modification and will be installed where the nick occurred. By programming the design of the pegRNA it is possible to introduce insertions, deletions and small changes in the DNA sequence.
Prime editing offers many advantages
Prime editing, like base editors, has the significant advantage of not requiring DSBs to perform gene editing. However, contrary to base editing, it offers the possibility to create a wider array of possible modifications. This is particularly important when trying to precisely edit, e.g., for gene correction, post-mitotic cells such as neurons or skeletal muscle cells.
For this approach to work, PNs must exploit homologous recombination (HR), one of the two major DNA repair pathways. However, in these cells, the HR pathway is either downregulated or completely shut down.
While this can be challenging for standard PNs, it is not an issue for prime editing as it does not rely on common DSB repair mechanisms. Additionally, researchers have shown that prime editing generates very few off targets compared to canonical CRISPR-Cas9 editing. This renders prime editing both a versatile and safe DSB-free technology particularly for cell types that would normally be difficult to edit.
Current prime editing applications
The potential of prime editing has not gone unnoticed and diverse research groups have started exploring its potential for clinical applications.
For example, the Olson lab at the University of Texas Southwestern has recently published a paper where prime editing was used in the context of Duchenne muscular dystrophy (DMD). In particular, prime editing was used to successfully restore the reading frame of the dystrophin (DMD) gene in human cardiomyocytes achieving up to 50% correction of the mutated Exon 52. The genetic correction was accompanied by the physiological amelioration of the arrhythmic defect seen in DMD patients with results close to what is observed in healthy cardiomyocytes.
Furthermore, prime editing has been also challenged in a variety of disease models by the group of Dr. Sabine Fuchs at Hamm-Lippstadt University of Applied Sciences in Germany who used this new technology – in combination with patient-derived organoids - to both precisely generate and correct intestinal and liver genetic conditions reporting high efficiencies and no genome-wide off-target effects.
A new era of effective and safe gene-editing therapies
When it comes to modifying the genetic sequence precisely, the standard CRISPR-Cas9 based applications usually yield mixed results that comprise a majority of indels while only a smaller fraction contains the desired modification. On the contrary, with prime editing, it is possible to install the changes desired with low or null undesired byproducts. Thus, prime editing is a new gene-editing tool, which in a DSB-free fashion enables precise genetic manipulation with high efficiency without compromising on safety. Recently, different research groups have started applying prime editing in different contexts, showing that this technology's hype is more than justified.
Although standard PNs approaches are being successfully involved in human clinical trials, there are still genetic conditions that may be beyond their reach because of limiting factors such as cell type (e.g. neurons, skeletal muscle cells, liver cells), the type of manipulation required or safety concerns. Prime editing becomes a new tool in the gene-editing arsenal with the promise answer certain clinical needs that otherwise would remain unmet.
Antonio Carusillo, PhD, currently employed at Vector BioPharma.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







