Prime Editing Restores Function of Genetic Defects in Patient-Derived Organoids
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Prime editing is a special flavour of the CRISPR/Cas9 genome editing technology, and it has recently become a new tool for Sabine Fuchs in her quest to develop new therapies for liver and metabolic disorders. Fuchs is a paediatrician at the Wilhelmina Children’s Hospital in Utrecht, the Netherlands who specialises in such genetic diseases, and for several years she has used 3D organoids as a tool in her studies. By expanding cells from the liver or intestines of patients, she has created an experimental avenue where such organoids can be used as disease models for toxicology studies, regenerative medicine, and most recently also gene therapy.
»Our results now open the avenue for human application, both for transplantation with ex vivo gene-corrected patient cells but – even more fascinating – for in vivo gene correction,« says Sabine Fuchs.
In October, Fuchs and colleagues published a paper in Nature Communications where they used patient-derived organoids to demonstrate that prime editing can be used for functional repair of disease-causing mutations. These included mutations leading to, for example, liver cancer and Wilson disease that causes storage of excess copper in various body tissues and is potentially lethal. And the researchers also showed that the procedure is safe in terms of unintended genetic modification.
Prime Editing is Efficient and Safe
Prime editing was first described in a Nature-paper in 2019 from the Liu-Lab at Harvard University in Cambridge, USA as a versatile and precise genome editing method that directly writes new genetic information into a specified DNA site. Read more in our interview with the first-author of that study, Andrew Anzalone.
Prime editing combines a prime editing guide RNA (pegRNA) with a catalytically impaired Cas9 endonuclease (Cas9-nickase) fused to an engineered reverse transcriptase. The pegRNA both directs the Cas9-nickase to nick the target DNA and instructs the reverse transcriptase to synthesise an edited DNA flap. The edited flap is then integrated by DNA repair mechanisms (called PE2), and this integration can be enhanced by also nicking the opposite strand (PE3) or doing so only after successful editing (PE3b).
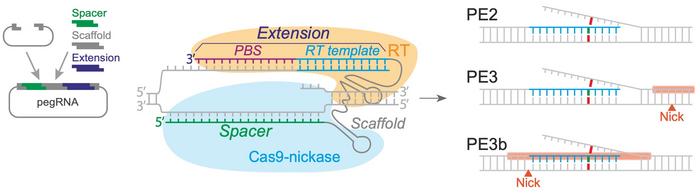
Prime editing has strengths and weaknesses compared to the other two most common CRISPR variants, namely base editing and Cas9-mediated homology-directed repair (HDR). Fuchs explains:
»Limitations to prime editing relate to the proximity of the mutation to a protospacer adjacent motif (PAM) site, and - as we noted in our initial experiments - low GC counts in the primer binding site were associated with inefficient prime editing. In terms of the type of mutation, prime editing can create all smaller mutations, including all nucleotide substitutions but also insertions and deletions (indels). However, both efficiency and safety of prime editing are significantly improved when compared to CRISPR-Cas9 HDR.«
The somewhat inefficient and error-prone Cas9-mediated HDR might still be the method of choice if safety is of no concern or if successfully corrected clones can be selected afterwards since it can perform virtually any desired edit. Base editing, on the other hand, relies on enzymatic conversion of bases and can only be achieved for four out of the twelve possible nucleotide substitutions. But if a mutation can be corrected by base editing, this is still the most efficient gene editing strategy.
Gene-Activation Leads to Formation of Organoids
Fuchs wanted to show functional correction of disease-causing mutations that could eventually be used to correcting cells and transplanting them back in the patient or even be used for in vivo gene editing. Accordingly, she chose to use prime editing because of its efficiency and safety as well as its versatility that allows most genetic disorders to be corrected.
“Typically, the first organoids can be observed after about a week of starting the patient-derived biopsies on the Wnt-activating organoid medium”Sabine Fuchs
She was already skilled at producing organoids using the protocols developed in the lab of Hans Clevers, who is also in Utrecht, by activating the Wnt signalling pathway in stem or progenitor cells taken from intestinal or liver tissues of patients. Wnt-activation leads to the proliferation of progenitor cells, which self-organise into three-dimensional structures termed organoids composed of cells bearing the disease-causing mutations.
»Typically, the first organoids can be observed after about a week of starting the patient-derived biopsies on the Wnt-activating organoid medium. In another one to two weeks, the culture contains enough organoids/cells to start gene-editing experiments,« says Fuchs.
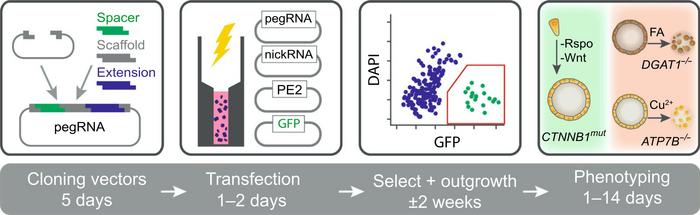
Selective Medium Reveals the Best pegRNAs
The intention was then to use prime editing for correction of these diseased organoids. But right at the start of the project, Fuchs and her colleagues encountered a major challenge.
»The biggest challenge in this study was that prime editing was still a very new technology and it was unknown which prime editing guide RNAs (pegRNAs) would be efficient. Therefore, we used functional assays to visualise successfully corrected clones quickly. This helped us to learn how to design efficient pegRNAs,« Fuchs recalls.
By functional assays, Fuchs means that the cell culture medium was adjusted to select for cells that had been effectively gene-edited. For example, the team wanted to screen for a pegRNA that could create a mutation in the β‐catenin gene (CTNNB1) that leads to proliferation independent of Wnt-stimuli and mimics a mechanism of liver cancer development. In that case, Wnt-protein was withdrawn from the culture medium, so only cells with the desired mutation in the CTNNB1 gene could proliferate.
Similarly, copper was added to the medium to select for correction of a mutation in the ATP7B gene that causes patients with Wilson disease to accumulate copper to toxic levels. In contrast, oleic acid was added to select for correction of a mutation in the DGAT1 gene from patients with congenital diarrhoea due to impaired lipid metabolism.
Gene Editing Efficiency Depends on pegRNA Design
Using this approach, the researchers could identify successful pegRNAs by observing a high number of surviving organoids and subsequently validate the correct edit by sequencing. For creating in-frame deletions in CTNNB1, striking differences up to a factor 50 were observed in editing efficiencies of different pegRNA designs. Similarly, large differences in pegRNA efficiencies were observed when prime editing was used to insert three missing nucleotides in the mutated DGAT1 gene.
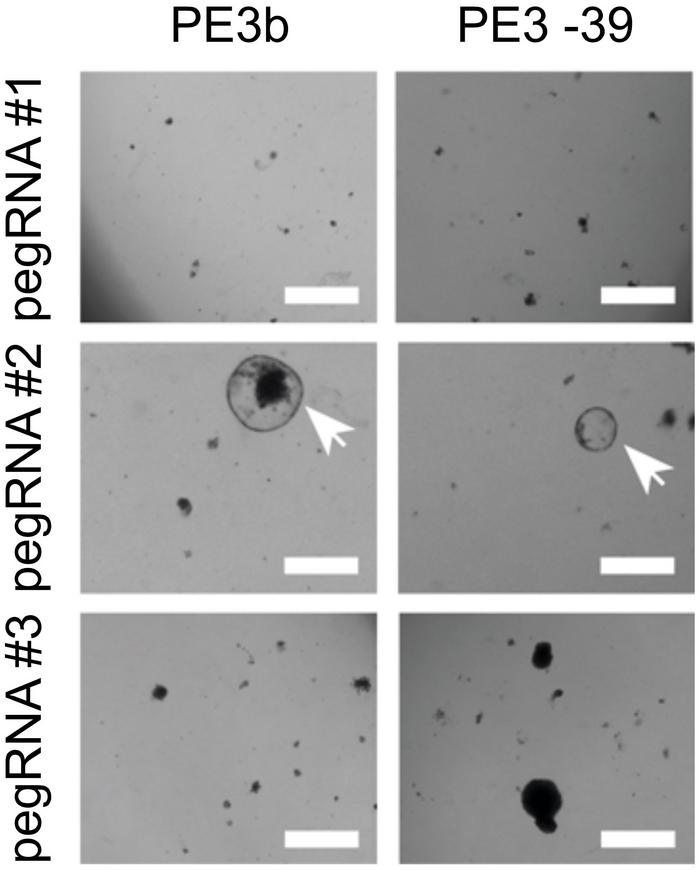
In an experiment without selection, prime editing of DGAT1 resulted in the correct edit in 21% of cells, while 8% of cells had indels and 71% contained no edit. These numbers were superior to Cas9-initiated HDR editing that led to only 6% of correctly edited cells, while 80% had indels and 14% contained no edit.
Not quite so successful was the attempt to correct a severe pathogenic G → A mutation in the ABCB11 gene that leads to bile salt export pump (BSEP) deficiency and liver failure. No correction was achieved using prime editing, while the same mutation was corrected in 20% of cells using base editing. These disappointing results were not entirely unexpected.
»Low GC counts in the primer binding site are associated with inefficient prime editing, and the two pegRNAs that we used in our efforts to correct ABCB11 had relatively low GC-content (27% and 33%, respectively). When we run our ABCB11-targeting pegRNAs in DeepPE - a webtool estimating prime editing efficiency for specific pegRNAs that became available after we published our results - we now find that they indeed have low predicted efficiency,« says Fuchs.
Prime Editing Can Be a Useful Therapy for Metabolic Disorders
The use of 3D organoids as disease models and prime editing for correction of genetic disorders both offers benefits and poses challenges, Fuchs explains:
»First of all, primary patient-derived cells have two crucial general advantages over immortal 2D cell lines: they better represent the tissue of origin, and they carry the complete genetic makeup of the patient that is studied. This makes them more suitable for performing functional assays that probe the disease phenotype. For gene editing specifically, patient derived organoids must be genetically stable. Immortalised cell-lines, on the other hand, have accumulated many genetic alterations due to selective pressure after serial passaging.«
Another promising result of the study was that prime editing did not induce any genome-wide off-target effects. This was demonstrated by whole-genome sequencing of both edited and unedited control clones, an effort that has previously not been conducted in human cells. But the disease model also poses challenges, says Fuchs:
»In this study, we first sorted single cells that had received the prime-editing machinery using transfection by electroporation. The organoids that grew from these single cells were genotyped and functionally investigated. However, established organoids are very difficult to transfect efficiently. One possibility to prime edit complete established organoids would be to put the prime editing machinery in a virus. This way, targeting efficiency would be high enough to correct a substantial percentage of cells.«
And targeting efficiency doesn’t need to be very high to have a therapeutic effect. Fuchs explains that in metabolic diseases, restoration of 5% of normal enzyme levels corrects most of the clinical phenotypes, especially when targeting the liver or hematopoietic stem cells. This means that correction of genetic effects by prime editing could eventually be a useful therapy for metabolic disorders like Wilson’s disease, phenylketonuria, glycogen storage disease, Crigler-Najjar syndrome, and other genetic diseases like hemophilia.
Link to the original article Nature Communications:
https://doi.org/10.1038/s41467-020-19136-7
Tags
ArticleNewsElectroporationCancerMetabolic disorderGene therapyOrganoidsOff-targetPrime editorsWilhelmina Children’s Hospital, Utrecht
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







