Revolving the Gene-Editing Landscape with Circular RNA-mediated Prime Editors
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Prime editing represents a significant advancement in gene-editing technology, allowing for precise DNA modifications without double-strand breaks (DSBs), thus minimising potential genomic instability. Traditional prime-editing systems, while effective, face challenges related to the delivery of their large components and the instability of linear prime-editing guide RNAs (pegRNAs). These linear pegRNAs are prone to degradation, and their structure can interfere with editing.
Chinese researchers have now developed a novel approach utilising circular RNA-mediated prime editors (CPEs) to overcome these obstacles, leveraging the increased stability and resilient nature of circular RNAs. The new system employs Cas12a, a smaller and potentially more precise alternative to Cas9, and offers improved editing efficiencies, multiplexing capabilities, and easier delivery. A selection of four alternative CPEs offers expanded applications in research, agriculture, and gene therapy.
»We've developed four distinct systems, each with unique editing efficiencies due to their engineering. Their primary advantage is efficiency, with some systems offering superior fidelity. Secondary is delivery, where split systems facilitate easier packaging,« explains PhD Kevin Zhao, co-first author of a recent paper in Nature Biotechnology presenting the new technology.
“Other researchers are exploring the use of Cas12a for prime editing, and we might not be the first to conceive the idea, but we are among the pioneers in implementing it”Caixia Gao
Zhao is the Chief Technical Officer of the Beijing-based gene-editing company Qi Biodesign, which he co-founded two years ago together with Caixia Gao. She is a professor and principal investigator at the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences in Beijing and senior author of the paper.
Slow nuclease allows for making a nickase
CRISPR Medicine News met Caixia Gao and Kevin Zhao when they visited Copenhagen earlier this month, and we discussed the new CPE system over lunch with Danish "smørrebrød". The system stands out for using circular RNAs that significantly influence editing efficiency and for introducing Cas12a in prime editing.
»Other researchers are exploring the use of Cas12a for prime editing, and we might not be the first to conceive the idea, but we are among the pioneers in implementing it. We chose Cas12a mainly for its small size,« says Caixia Gao.
This approach marks a significant departure from the reliance on the larger Cas9 system. The CRISPR-Cas12a (or Cpf1) family is extensive, with many small proteins like Cas12b (C2c1) and Cas12i (CasPhi). These smaller proteins could potentially be utilised in human and plant cells, making this platform's demonstration pivotal for future research, without the limitations of Cas9.

However, it was a challenge to use Cas12a for prime editing, since prime editing requires nickase activity to avoid the DSB formation. Cas9, known for its dual nuclease domains (RuvC and HNH), contrasts with Cas12's single RuvC domain, which traditionally offers a binary operational mode - either fully active or inactive. The Chinese research team eventually discovered mutations in Cas12a that modify its activity. Kevin Zhao explains:
»Normally, Cas12a cleaves both DNA strands, starting with one and then the other. We identified a mutation that delays the second cut significantly, which is ideal for our purposes. By making the first cut and delaying the second, we achieve the desired nicking effect for prime editing. This mutation facilitates what we call a pseudo-nickase or a kinetic nickase.«
Circular RNA means high prime editing efficiency
Another prime editing-related issue with Cas12a is its RNA processing capability, which can undesirably truncate extended guide RNAs. This challenge was circumvented by circularising the RNA, showcasing a unique solution tailored for Cas12a, which is not required for Cas9.
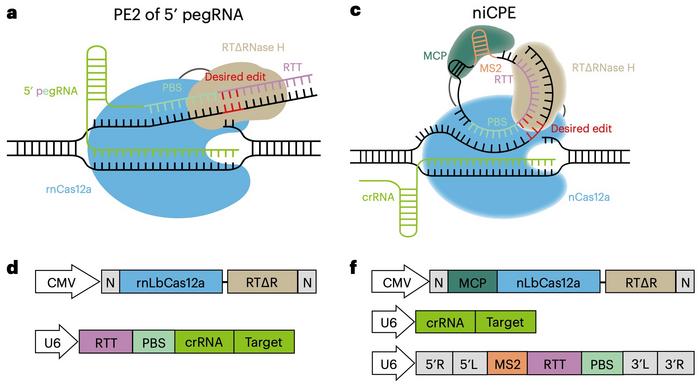
Typically, the pegRNA includes a crRNA guide, a reverse transcriptase template (RTT) and a primer binding site (PBS); this results in a long, linear molecule that is unstable and susceptible to degradation by nucleases. However, the pegRNA can be split into a linear crRNA and a circular RNA containing the RTT and PBS that lacks dissociative ends and thus is more stable (see Figure 2).
The first iteration of a circular RNA-mediated prime editor that the scientists tried with the kinetic nickase Cas12a was a nickase-dependent CPE (niCPE). Targeting eight different sites in HEK293T cells yielded editing efficiencies ranging from 1.56% to 10.13%. Moreover, the niCPE editor could create base insertions (up to 21 bp), deletions (up to 48 bp), and substitutions (up to 10 bp) efficiently, with efficiencies ranging from 2.41% to 24.06%.
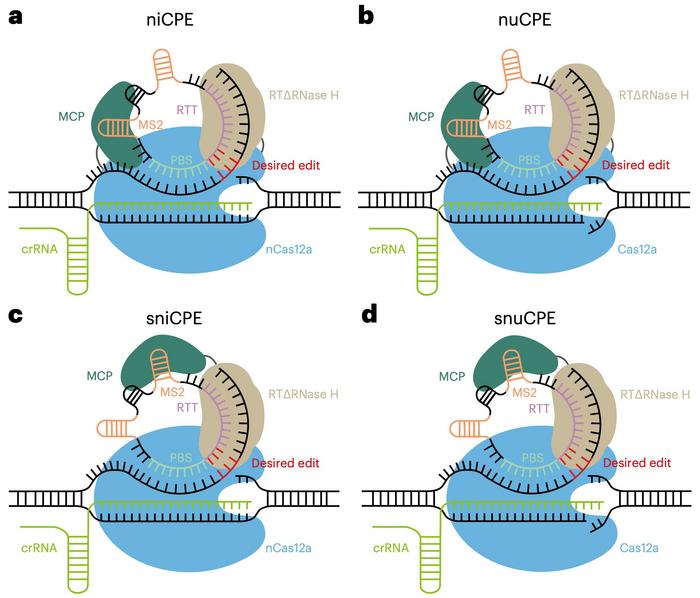
The team designed three more CPE variants: nuCPE, sniCPE, and snuCPE (see Figure 3). nuCPE utilises the normal Cas12a nuclease instead of the Cas12a kinetic nickase, and the "s" prefixes denote split constructs that express Cas12a and the reverse transcriptase complex separately.
»Prime editors are large, making it challenging to package a complete editor within a single AAV. By dividing the components, we facilitate easier packaging and delivery. The split systems generally provide better delivery options and editing efficiencies,« says Kevin Zhao.
Four CPEs allow for flexibility
When tested for precise editing in HEK293T cells, the performance of the four variants differed according to the target site. In most cases, the niCPE and sniCPE nickase variants outperformed the others, and the latter yielded up to 40.75% editing efficiency. The efficiencies of niCPE and nuCPE reached 4.13%-14.93% and 1.14%-10.42%, respectively. The nuclease versions, nuCPE and snuCPE, induced more indels than the nickase versions, niCPE and sniCPE. Generally, the CPE systems enabled efficient and specific prime editing with hardly any off-target editing.
The four CPEs allow for balancing desired editing activity and indel formation, and the system choice depends on the application's requirements. The split systems offer enhanced delivery options and potentially greater editing efficiencies for human therapeutic applications by fitting within the AAV capacity constraints. The nickase systems are favoured in medical applications where minimising indel formation is critical. In contrast, the nuclease variants can be helpful for plant applications where the consequences of indel formation can be mitigated through screening processes.
Another advantage of the new CPE system is that it gives more flexibility in designing prime editing experiments. While Cas9 requires a G-rich NGG protospacer-adjacent motif (PAM), Cas12a recognises a T-rich TTTV PAM (where V can be A, C, or G).
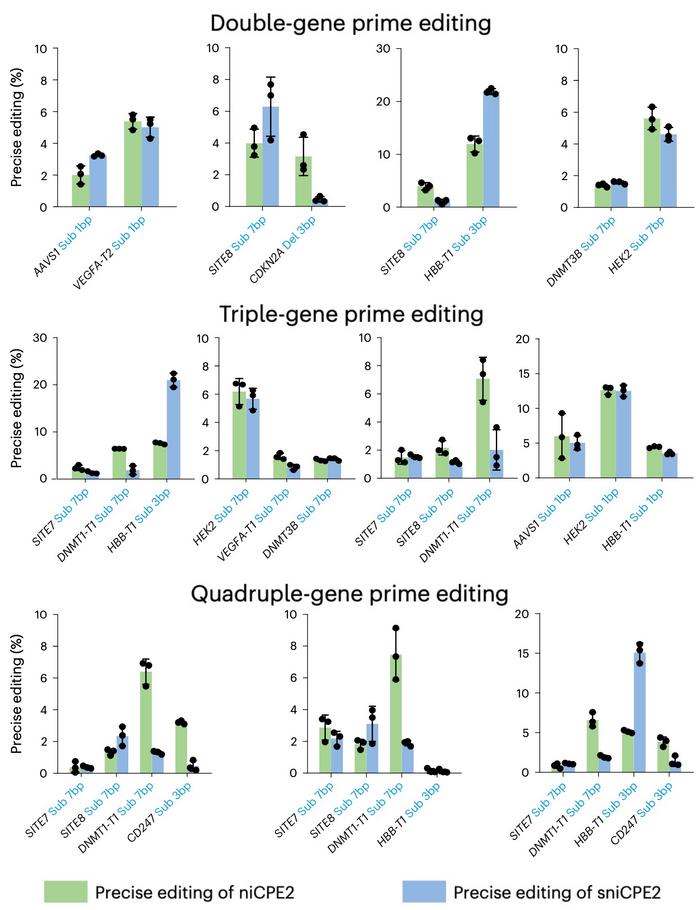
Moreover, the niCPEs and sniCPEs were shown to prime edit multiple genes simultaneously and very efficiently, when two or more crRNAs were included in the construct (see Figure 4). Editing efficiencies varied depending on the CPE system, the target and the combination of targets, but they were generally high. For example, prime editing frequencies at the HBB-T1 site with sniCPE2 were 21.87% in double-gene prime editing, 20.99% in triple-gene prime editing and 15.08% in quadruple-gene prime editing.
Both humans and plants will benefit
Qi Biodesign, where the new Cas12a and circular RNA system for prime editing in human cells is developed, focuses on gene editing in plants for agricultural use. There was, however, a good reason for using human cells rather than plant cells in these experiments, Kevin Zhao explains:
»While there are various methods to produce circular RNA, the most prevalent one involves using mammalian cell machinery. We initially employed this machinery to establish a proof of concept in mammalian cells with the CRISPR-associated protein Cas12a. We are now working on adapting and optimising this for plant cells, including developing methods to produce circular RNAs in vitro for delivery into plants.«
“We have already engaged in clinical collaborations, such as with a CAR-T company in China, to explore the application of our technologies in their projects”Kevin Zhao
He sees great potential in prime editing and believes the advances will likely lead to clinical trials soon. Though Qi Biodesign, which currently has around 70 employees, mainly works with plants, they closely follow gene editing developments in the human therapeutic field.
»Our focus is on plant applications, but we are open to collaborating with companies working on human therapeutics. Our platform, including base editors and prime editors, is available for licensing and partnerships. We have already engaged in clinical collaborations, such as with a CAR-T company in China, to explore the application of our technologies in their projects,« concludes Kevin Zhao.
Link to the original article in Nature Biotechnology:
Prime editing using CRISPR-Cas12a and circular RNAs in human cells
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.