Spatio-Temporal Control of CRISPR Editing With CRISPRoff

A team of scientists from Silicon Valley-based genome engineering company Synthego recently developed CRISPRoff, a revolutionary light-controllable CRISPR tool that makes it possible and easy to control temporal and spatial editing, while also increasing the ratio of on: off target editing events.
Robert Deans, PhD and Chief Scientific Officer at Synthego, highlights the significance of CRISPRoff: »We wanted to invent a technology that could control temporal and spatial CRISPR editing in cells, that was simple, easy to control, and universally applicable, and this is exactly what we’ve gotten with CRISPRoff«.
One of the obvious advantages of using an optically-controlled system such as CRISPRoff is the potential to study precise spatial control of CRISPR editing. The possibility to study the effects of gene editing in specific tissue regions is a game-changer in many areas of research and medicine.
CRISPRoff Guide RNAs
Despite advances in precision, targeting range and applicability of CRISPR to therapeutics in recent years, tools to temporally and spatially control CRISPR editing are lagging behind. Without this control, editing can continue freely in cells until the machinery i.e. Cas endonuclease and single guide RNAs (sgRNA) degrades through cellular processes. Uncontrolled CRISPR editing carries the risk of off-target edits that may result from prolonged or uncontrolled Cas activity.
Recognising that the sgRNA is the most easily programmable component of the CRISPR system, Robert Deans and his team set out to synthesise sgRNAs that could be deactivated in the presence of light.
Jared Carlson-Stevermer, lead author on the study that was recently published in Nature Communications explains the principle behind CRISPRoff:
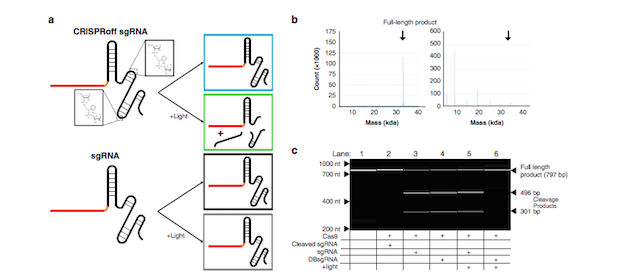
»We added o-nitrobenzyl groups, which are sensitive to cleavage when exposed to ultraviolet light, at two optimised locations on the sgRNA. This dual breakage sgRNA (DBsgRNA) is prone to fragmentation upon illumination with light of a specific wavelength. We rationalised that once the DBsgRNA is transfected in cells with Cas9 in RNP format, it would edit normally, but exposure to light would result in fragmentation of the sgRNAs and halt further CRISPR editing«.
The team synthesised the modified DBsgRNAs, known as CRISPRoffs using solid phase synthesis, which allowed them to develop a universal sgRNA template that is photocleavable and still retains gene-editing capacity comparable to their standard sgRNA counterparts.
They first generated a variety of CRISPRoff molecules in which fixed positions along the backbone were replaced by photocleavable residues, and evaluated the fragmentation patterns of the modified guides upon exposure to broad-spectrum light. The team then selected promising CRISPRoffs to form RNP complexes that were assessed for editing activity in vitro in human cell lines.
These experiments revealed that it was possible to replace two sites on the sgRNA backbone with photocleavable residues and still retain good editing efficiency.
Universal Control Across Human Cells Lines and Genomic Targets
With the groundwork in place, the team then found the system to be efficient in several human cell lines and most of the 26 genomic targets tested. Indels (off-target insertions and deletions) were observed at low and comparable rates across all sgRNAs and CRISPRoffs tested.
In addition, they observed that photocleavage at either residue deactivates the CRISPRoff, increasing the likelihood that at least one site is cleaved upon illumination and thus strengthening the safety of CRISPRoff.
Improved On: Off Target Event Ratios
While testing their panel of CRISPRoffs against genomic targets, the team included a known cytotoxic sgRNA and corresponding CRISPRoff, and remarkably they observed greater cell survival rates when this CRISPRoff was exposed to UV light. This hinted at an increase in the ratio of on: off target events, which the team was able to verify in subsequent experiments, revealing that controlling DSB formation with the CRISPRoff system reduces off-target CRISPR editing.
On the topic of safety, Jared Carlson-Stevermer also told us: »Our research was not focused on explicitly testing toxicity in vivo, but we can confirm that we did not observe cellular toxicity in our experiments.«
While most of the CRISPRoffs responded to light, the effects of editing efficiency was variable. The team found some evidence that this could be related to individual editing kinetics at each target site.
Jared Carlson-Stevermer shared some additional thoughts on this observation: »Editing efficiency can be different across cell lines and target sites for a variety of reasons, such as local chromatin context, cell cycle state, and the related, predominant repair pathway. This is an interesting area for further study, as we continue to invent fully-controlled editing mechanisms.«
Spatial Patterning With CRISPRoff
The team explored the potential for spatial control with CRISPRoff using a GFP-expressing cell line as a proof of concept and found that it was possible to selectively disrupt GFP expression and thus create a visual patterning effect in distinct regions of a thin layer of cells.
»Our data demonstrates that CRISPRoff is effective and can be immediately useful across multiple genomic targets in multiple cell lines with the potential to optimise on-target editing events and the ability to spatially pattern cells in vitro«, said Robert Deans.
Readily and Easily Integrable into CRISPR RNP Workflows
Others researchers have explored means to control CRISPR editing inside cells. The most widely used strategies involve controlling Cas expression through the use of doxycycline-inducible on-off gene expression switches, or controlling sgRNA expression, e.g., by using a Cre-Lox system that allows controlled activation and deactivation of target genes.
Other approaches include modulating the activity of sgRNAs through ligand binding, anti-CRISPR proteins and the use of partial Cas endonucleases that dimerise and become activated in response to certain stimuli, e.g., blue light. Although these approaches have been used with some success, they suffer a common drawback. They all require the delivery of an additional CRISPR component to the cell besides the RNP or delivery vectors containing the Cas, sgRNA and repair template as necessary. The need to deliver additional material exasperates the already existing challenge of delivering CRISPR components to cells as well as adding an extra processing step that may impact the overall success of the workflow.
Robert Deans explains how CRISPRoff stands out from the other approaches: »CRISPRoff is a programmable CRISPRoff that can be integrated into existing CRISPR workflows without the additional need for optimisation or special experiments. While we demonstrated proof-of-concept using Cas9 in this study, the basic idea can be expanded to other nucleases, and the photosensitive groups are also amenable to variations«, who proceeds to remind us that CRISPRoff sgRNAs are in fact identical to standard sgRNAs except for the replacement of two specific nucleotides with o-nitrobenzyl residues.
»We believe that CRISPRoff will work as well as conventional sgRNAs across all genomic targets«, said Deans.
One potential limitation of CRISPoff, however, is that the chemical modifications required for CRISPRoff photocleavage can only be added synthetically during CRISPRoff synthesis. Therefore, the CRISPRoffs are not compatible with CRISPR workflows that rely on in vitro / in vivo sgRNA transcription.
Therapeutic Potential
While it’s exciting to speculate about a place for CRISPRoff in the development of in vivo therapies in the future, Jared Carlson-Stevermer tells us that this is not the main focus at present: »CRISPRoff is currently focused on in vitro applications, due to the low penetrance of UV light through tissues«.
However, he quickly ensures us that the current in vitro focus does not preclude the therapeutic potential of CRISPRoff.
»We see the CRISPRoff system having real value for cell therapies that rely on ex vivo editing strategies«, said Jared Carlson-Stevermer.
A New Tool in the CRISPR Toolbox
Temporal and spatial control is not the only attractive feature of CRISPRoff however, as Robert Deans explains: »We believe that CRISPRoff can be used to build the highest safety margin for gene editing based on the increased on: off targeting rates observed in our study, which in turn can provide a clear and rapid lot release strategy. This will allow accelerated movement into the translational space and helps fulfill our vision to make these new ex vivo edited cell therapies rapidly accessible«.
What’s next for CRISPRoff then? Robert Deans takes the last word: »We plan to continue to design highly-controlled CRISPR-based genome engineering platforms and products, and a future where clinical-grade sgRNA amenable to easily being switched off is available off-the-shelf is not far away«.
Link to original article in Nature Communications:
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







