Switchable CRISPR-Cas9 guide RNAs for sequential gene editing

It has long been a dream to study the multiple genetic steps of complex biology in cells. Now researchers have leveraged the remarkable capacity of CRISPR-Cas9 gene editing to develop a new technique that allows for the programming of sequential editing over time.
»The ability to preprogram the sequential activation of Cas9 at multiple sites introduces a new tool for biological research and genetic engineering,« says Dr Bradley Merrill. He led the new study at the University of Illinois in Chicago.
»In biological processes, time is a critical component, and the order of genetic events can vary the outcome — especially during development and disease progression. Our system allows researchers to precisely preorder multiple genetic events in a cell to investigate these time-sensitive processes,« Merill adds.
Recording the history of genetic events
For the past 15 years, Merrill’s lab has been studying embryonic development, and his group has extensively used genetic tools to manipulate the genes of their interest. With the advancement in CRISPR-mediated gene-editing technologies, Dr Merrill saw an opportunity to develop a tool that would record the history of genetic events in the cell in its genomic DNA. Thus, they worked on developing a system that would trigger stepwise genetic events in a preordered way to follow the cell lineage and differentiate between mother, daughter, and granddaughter cells.
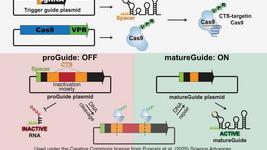
In the recent paper published in Molecular Cell, his group has developed a new method that allows researchers to enact multiple CRISPR-Cas9 activities together in a preordered way. The system relies on a set of inactive guide RNA-like components (proGuides), each of which is converted to an active state by the action of another activated guide RNA.
Engineering the proGuide
The CRISPR-Cas9 system uses single-guide RNA (sgRNA) to direct Cas9 to initiate a cut in the desired location.
Cas9 is a very efficient enzyme, and it can simultaneously trigger multiple genetic events in a cell, making it difficult to sequentially order multiple genetic events.
Researchers in other labs have used chemicals to control genome editing, but the availability of such chemicals limits these methods. Another limitation of chemical-mediated sequential gene editing is that the second genetic event is not dependent on the first event, which generates a heterogeneous mixture of cells.
In other words, the first and the second event might take place in two different cells creating a mixture of cells with various genetic changes since the genetic events are regulated by different chemicals independent of each other.
To create a system that allows cells to progress through a series of instructions in sequential order, the researchers inactivated guide RNAs (proGuide) by inserting a ribozyme-encoding DNA with stem structures on either side.
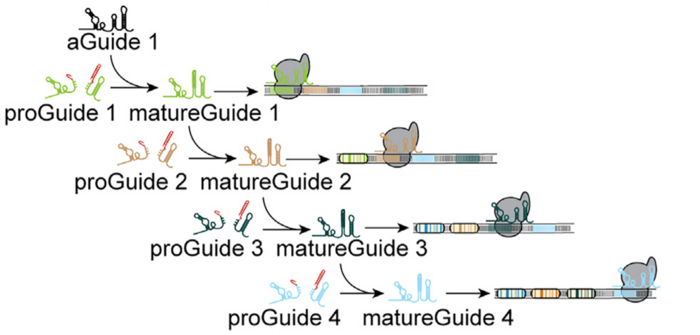
Activation of proGuide requires removal of ribozyme-encoding DNA via cleavage on the stem structure flanking its sides. The researchers used an active guide (aGuide) to activate the first proGuide producing matureGuide1. The matureGuide1 then acts as an active guide for proGude2, which then makes matureGuide2. Thus, regulating the sequential activation of guide RNAs based on previous genetic event.
»The advantage of this system is that you don’t have to worry about cell heterogeneity. Since, in this system, the second event won’t happen unless the first event takes place. So, it allows researchers to study biological events in a time-dependent manner,« says Bradley Merrill.
proGuide validation
To validate the concept, the researchers inserted ribozyme-encoding DNA and the flanking stem structures at two different sites (hairpin1 or tetraloop) of sgRNA targeting EGFP (fluorescent protein) in 4T1 cell. The ribozyme-encoding DNA effectively disrupted the sgRNA and prevented its spCas9-related activity with very low leakiness (0.36%).
The activating guide (aGuide) to the cell triggered proGuide and disrupted the GFP signal in 40% of the hairpin and 48% of tetraloop variant proGuide.
The proGuide triggered genome editing led to ribozyme disruption in 36.2% of the cases and showed slower gene editing kinetics. It suggests that the removal of the ribozyme is the efficiency-limiting step in the process.
»With the current system, we are still far from tracing genetic events among cell progenies. Still, we have realized a lot of other potentials for proGuide based sequential gene manipulation,« says Bradley Merrill.
As an example, he points to the development of complex genetic disease like cancer requires multiple genetic events. A set of genes get activated, and others get inactivated to transform a healthy cell into a malignant cell.
»The proGuide based system enables researchers to manipulate genes in a sequential manner, which will allow them to determine the sequence of events a cancer cell undergoes from acquiring oncogenic mutation to gaining a metastatic potential,« says Bradley Merrill.
Link to the original article in Molecular Cell: Sequential Activation of Guide RNAs to Enable Successive CRISPR-Cas9 Activities
Shishir Pant is a cancer biologist and a scientific writer based in Helsinki, Finland.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.