Tiny but Mighty: How Researchers Found a New Game-Changing CRISPR Tool
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Since its discovery as an ancient prokaryotic immune system, CRISPR-Cas has been harnessed as a programmable and precise gene editing tool within research and clinical development in labs worldwide. However, despite massive advances in gene editing efficiency and precision, cellular delivery of the Cas endonucleases and guide RNAs - the machinery that is necessary for gene editing - remains a key challenge, mainly because of the large size of the Cas proteins.
Researchers at the University of California, Berkeley have now characterised CasΦ, a new member of the CRISPR-Cas arsenal that originates from the largest known bacteriophages. CasΦ targets and cleaves bacterial, human and plant DNA and is only about half the size of the CRISPR workhorse Cas9, making it a very attractive addition to the gene editing toolbox.
I caught up with Basem Al-Shayeb and Patrick Pausch PhD, graduate student and postdoctoral scholar respectively, from Jennifer Doudna’s Lab at the University of California (UC), Berkeley, and joint first authors on the work that was recently published in Science.
Tiny CRISPR Systems in Huge Bacteriophages
Basem Al-Shayeb was inspired by the findings in Kim Seed’s Lab at UC Berkeley, which identified the first phage-encoded CRISPR-Cas system in the pathogenic bacterium Vibrio cholera.
»It seemed like this phage in V. cholera picked up a CRISPR system to annihilate a host defense system and I thought that was really fascinating. I started looking into CRISPR systems on phages because this phenomenon was not further explored in the viral universe since the initial discovery. It was actually thought to be a rare occurrence. I wanted to look at what other CRISPR systems might be out there in phages, and what they could be doing«, said Basem Al-Shayeb about the discovery path to CasΦ.
Basem Al-Shayeb didn’t just look for CRISPR systems in already identified phages, though. Last year, as a joint graduate student in the lab of Jill Banfield, also at UC Berkeley, Basem co-first-authored a Nature paper describing new clades of huge bacteriophages, discovered through metagenomics studies of diverse ecosystems across the Earth. While digging into these huge metagenomics datasets, Basem made another surprising discovery. He explains:
»I kept finding CRISPR systems in the long unknown fragments of DNA that turned out to be huge bacteriophage genomes. Coincidentally, the phages that seemed to harbour new CRISPR systems and the phages that were huge were the same. We found the previously discovered bacterial CRISPR systems on phages, but we also found new systems that were very small.«
CasΦ is Discovered
Although they found several CRISPR systems in phage genomes, Basem Al-Shayeb and colleagues focused on one in particular from the Biggiephage clade that they decided to call CasΦ (Figure 1; bacteriophages are often abbreviated as Φ).
»CasΦ was the most interesting system to us because it was the only one that we found exclusively in phage DNA. That made us think there might be something special about this protein, that it might do something that the others can't.« explained Basem Al-Shayeb.
CasΦ was indeed peculiar.
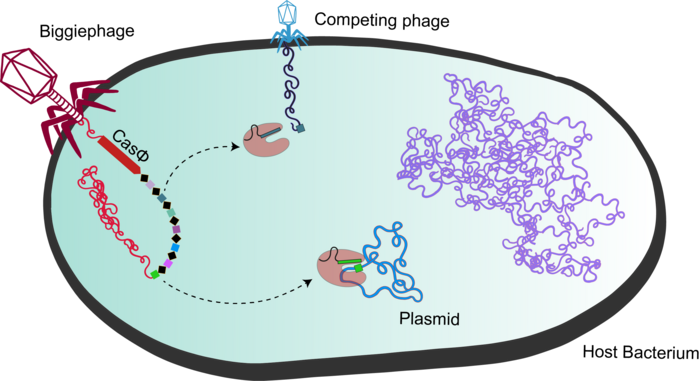
Bacterial CRISPR-Cas systems such as CRISPR-Cas9 rely on the Cas1, Cas2 and Cas4 enzymes to acquire new spacers in the bacterial genome. Spacers are short genome fragments from previous phage infections that are stored in bacterial genomes as a record of infection, and serve as an immunological memory. The loci that encode CasΦ appeared to encode just that one protein, and there was no sign of the enzymes required to acquire new spacers.
»This led us to ask whether the CasΦ orthologs we discovered were real Cas proteins that use guide RNAs to cut target DNA, or an evolutionary intermediate between the phage ancestor and the CRISPR-Cas systems we know from bacteria«, said Basem Al-Shayeb.
Casφ Is Functional, Compact and Cleaves Double-Stranded DNA
On a sequence level, CasΦ orthologs are only remotely similar to bacterial Cas endonucleases, and they are about half the size of the well-characterised Cas9 and Cas12a enzymes with a molecular weight in the range of 70-80 kDa.
These observations raised the possibility that CasΦ might have limited functionality, for example the ability to only cut single-stranded DNA, as is seen for other compact Cas proteins identified to date, for example Cas14, which was discovered in the Doudna Lab in 2018.
The researchers devised a simple yet clever plasmid rescue assay to check whether or not CasΦ could recognise and cut target double-stranded DNA (dsDNA) at all. Patrick Pausch explains the setup: »We use two plasmids. One encodes the CasΦ enzyme and a CRISPR RNA (crRNA) that is complementary to a sequence on the second plasmid, which is essentially the target. If CasΦ recognises the target plasmid, it will cleave it.«
CRISPR-Cas systems target DNA sequences that lie either side of a 2- to 5- bp protospacer adjacent motif (PAM) for self versus nonself discrimination.
The researchers investigated CasΦ’s PAM dependence by making a library of target plasmids with short random sequences next to crRNA-complementary target sites that might function as potential PAMs. By mixing the effector plasmid with the target plasmid library and reisolating surviving target plasmids from transformed E. coli, the researchers could identify via sequencing which plasmids had been depleted (by CasΦ cleavage) and thereby infer the PAM specificity of CasΦ.
With this setup, the researchers studied 3 divergent CasΦ orthologs from the metagenomics assemblies and found that all 3 were capable of crRNA-guided dsDNA cleavage and that this activity depended on a minimal T-rich PAM sequence. Similarly to Cas12a, CasΦ cleaves dsDNA leaving staggered 5’ overhangs of 8-12 nucleotides in length.
CasΦ-CRISPR is a Hyper Compact Efficient Gene Editing System
Using RNA sequencing to track RNA expression from the CasΦ loci in bacterial expression vectors, the researchers saw that these loci could express casΦ and a reduced CRISPR array (containing only a few spacer sequences), but there was no evidence for expression of transactivating CRISPR RNAs (tracrRNA) or other non-coding RNAs that are sometimes expressed in CRISPR-Cas systems.
These findings hinted that CasΦ was much more compact than bacterial CRISPR-Cas systems and corroborated what the researchers found in the genome analysis and later in the plasmid rescue assay, where CasΦ and a crRNA were sufficient for maturation of the guide RNA for target recognition and cleavage. This level of compactness has never been seen for a CRISPR-Cas system before, making CasΦ the smallest functional CRISPR system identified so far.
Further experiments revealed that CasΦ targets and cleaves dsDNA in bacterial cells, in human embryonic kidney cells and in cells of the model plant Arabidopsis thaliana. Because CasΦ recognises minimal T-rich PAM sequences, it is anticipated to target a broader range of DNA sequences than Cas9.
A Versatile New Tool in the Human and Plant CRISPR Toolbox
The researchers describe their work so far as a proof of concept, and they are already working to optimise many aspects of CasΦ to make it an even more attractive gene editing tool. Leaning on the huge body of knowledge already gained in the Cas9 and Cas12a fields, they have already seen improvements in targeting efficiency and they will push this further.
Given the very small size of CasΦ and its highly compact nature, it is obvious to explore cellular delivery with adeno-associated virus (AAV) delivery vectors for therapeutic applications. One other therapeutically-relevant advantage of CasΦ is that because it originates from phages, which do not infect humans, the risk of rejection of CasΦ-based therapies might be lower than with bacterial-derived Cas9-based therapies, given that anti-Cas9 antibodies have already been found in a number of individuals.
Besides the obvious attraction of CasΦ for therapeutic gene editing, this tiny CRISPR system may also be very useful in agricultural applications. Unlike Cas9, which is most efficient at 37 degrees Celsius, CasΦ is not temperature-sensitive, with good activity at temperatures that support plant growth, and it has already been shown to cut dsDNA in plant cells. The small size of CasΦ is also advantageous for CRISPR in plants where there are limitations to the amount of material that can be packaged into plant-compatible delivery vectors.

There are many reasons to be excited about CasΦ but only time will tell exactly where it fits into the CRISPR toolbox. In the meantime, its discovery reminds us of the power of basic research. Basem Al-Shayeb has the last word:
»A lot of people tend to think of biology as a place to find a new tool that we can use, but the really useful and groundbreaking discoveries tend to come more from an interest in basic biology. When we were looking at huge bacteriophages we weren't expecting to find something like this. And when people were looking at CRISPR in the very beginning, they weren't thinking about it as a way to cure genetic diseases? They just saw weird repeats in weird archaeal genomes.«
Link to original article in Science:
CRISPR-CasΦ from huge phages is a hypercompact genome editor.
The Banfield and Doudna Labs
Jill Banfield is a Professor in the Departments of Earth and Planetary Science and Environmental Science, Policy, and Management at UC Berkeley. Her lab studies how microorganisms shape, and are shaped by, their natural environments. The group studies various aspects of microbial communities, primarily using cultivation-independent approaches such as genomics (metagenomics) and community proteomics.
Jennifer Doudna is the Li Ka Shing Chancellor’s Chair and a Professor in the Departments of Chemistry and of Molecular and Cell Biology at UC Berkeley, as well as an Investigator of the Howard Hughes Medical Institute. She was one of the discoverers of CRISPR-Cas9 and today her lab continues to explore and develop CRISPR systems and other RNA-guided mechanisms of gene regulation.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.








