Truncated guide RNAs Protect Off-Target Sites from CRISPR-Cas9 Editing
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

You have probably heard about 'dead Cas9' but now it is time to get used to 'dead RNA' as well. A dead RNA (or simply a dRNA) is a truncated gRNA that attaches to a functional Cas9 and turns it into a nonfunctional Cas9. In this way, dRNAs can control not only where Cas9 is going to bind the DNA but also whether it’s going to cut the DNA or not.
Dead RNAs have now been used to direct Cas9 to predicted off-target sites where the inactive complex binds and thereby shields the site from unwanted cleavage by the functional gRNA-Cas9 complex. This novel approach to increase the efficiency and safety of CRISPR was presented in a paper published in Nature Communications in June by Douglas Fowler, who is Associate Professor at University of Washington in Seattle, USA.
»Dead RNA off-target suppression - or simply dOTS - has the potential to become a regularly used tool in the Cas9 toolbox,« says Fowler. He envisions that the ease of designing, screening, and implementing dRNAs into existing Cas9 workflows is likely to facilitate the widespread use of dOTS in the future.
The rationale behind the new approach is that gRNAs are usually required to be 17 nucleotides or longer to facilitate cleavage by Cas9. If the gRNA is 16 nucleotides or shorter, it can still direct Cas9 to a matching sequence, but cutting is hampered.
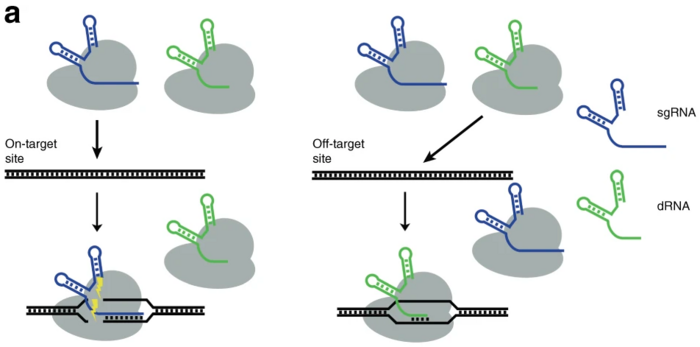
Competition increases efficiency
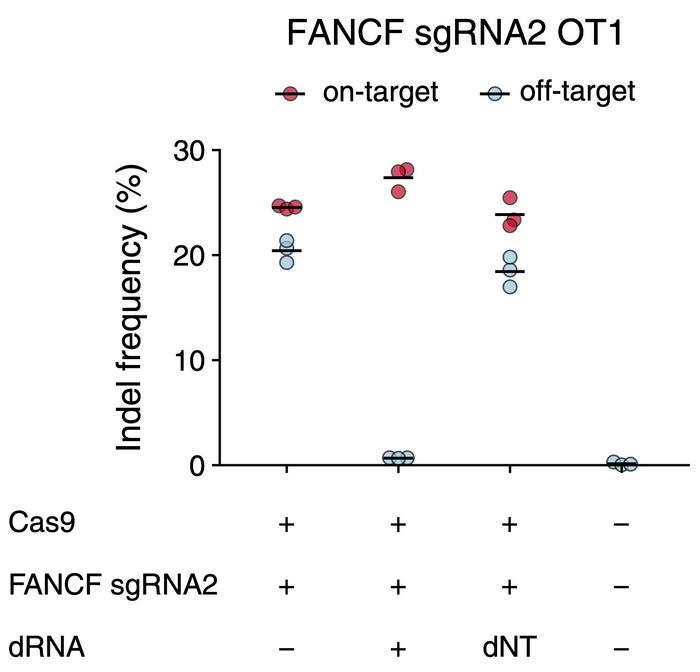
Fowler and colleagues tested the approach by co-transfecting human cells with a mixture of plasmids encoding Cas9, gRNA targeting the FANCF (Fanconi anemia group F) gene, and one of four dRNAs designed to target a known off-target site, OT1. In the absence of dRNA, on-target editing was only slightly higher than off-target editing with indel frequencies of 24 % and 20 %, respectively. Co-transfection with the best performing dRNA, however, decreased off-target editing to only 1 % while on-target editing increased to 27 %.
In other words, the presence of dRNA increased the on-target vs. off-target specificity ratio 30-fold and thereby highlighted the potential of the dOTS strategy. To evaluate if these observations were uniquely associated with the FANCF-OT1 combination, the scientists repeated the experiment with 19 other on-target/off-target pairs.
In 17 of these cases, the specificity ratio increased on average 10-fold and in all but 2 cases, the on-target editing frequency increased as well. Importantly, Fowler and his co-workers also checked whether the dRNA-Cas9 complex leads to detectable cleavage anywhere else in the genome. They found that not to be the case.
»In our work, we see minimal evidence that a dRNA-Cas9 designed to block an off-target site impedes the sgRNA-Cas9 complex from cleaving the on-target site,« Fowler points out. So while the dRNAs and gRNAs compete for the same sequences, the nonfunctional dRNA-Cas9 complex doesn't bind to and block on-target sites but instead promotes the functional gRNA-Cas9 complex to do so.
»The reasoning here is two-fold,« Fowler explains: »First, the dRNA is imperfectly complementary to the on-target site and in many cases has little complementarity to the on-target site. Instead, the higher affinity gRNA outcompetes the dRNA for the on-target site. Secondly, the truncated dRNA has fewer base pairs to bind to the DNA on-target site, meaning it is likely even less tolerant of mismatches than a gRNA.«
dOTS facilitates scarless HDR
Fowler and his colleagues don't have a general guide to design dRNAs for a specific purpose, but there seems to be a few rules to keep in mind. Firstly, the dRNA must be no longer than 16 nucleotides, and secondly, it should contain at least one mismatch to the on-target site. When these simple rules are adhered to, the researchers usually only needed to screen 4-8 dRNAs to find an effective one for the majority of the sites tested.
There were, however, a few cases where the scientists could not easily find a dRNA that effectively competed with a specific gRNA. Fowler believes that PAM sequences are involved in this.
»The primary limitation to identifying an effective dRNA for dOTS is the availability of off-target proximal PAMs. The use of engineered Cas9 variants with more permissive PAM requirements would increase the number of candidate dRNAs at difficult to suppress sites,« he suggests.
Aside from reducing editing at off-target sites, dOTS can also be used to facilitate scarless homology-directed repair (HDR) in a process Fowler has dubbed dRNA ReCutting Suppression or dReCS. In many cases, the target sequence or PAM is not substantially altered after HDR and thus allows Cas9 to recut the repaired site. Fowler found, however, that dRNAs could be designed to block repaired sites from recutting and thereby double HDR efficiency.
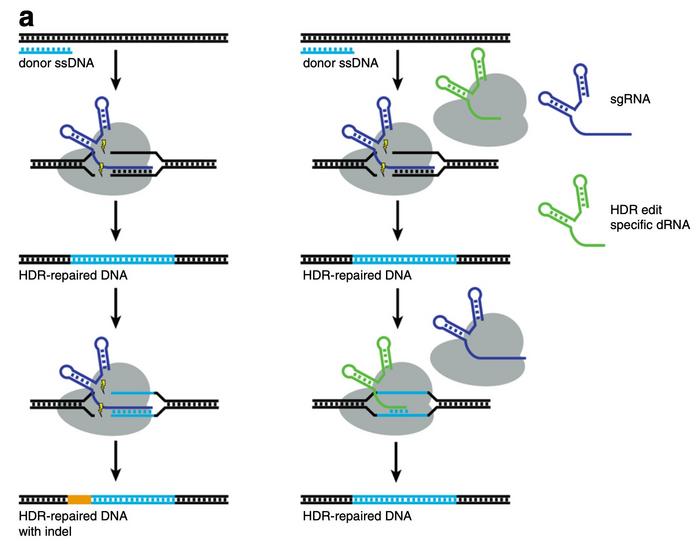
Further validation is needed for clinical use
The team at the University of Washington found even further uses of dOTS. For instance, dRNAs could be multiplexed to shield three off-target sites of a single gRNA at the same time or to suppress off-targets of two distinct gRNAs simultaneously. Moreover, dOTS was shown to synergise well with existing tools such as high-fidelity Cas9 variants to improve on-target specificity.
It seems obvious to use dOTS and dRNAs in the clinical setting where the unwanted and unpredictable effects of off-target editing are of major concern. But Fowler stresses the need to fully validate that the dRNA has no editing capability at the gRNA off-target site or any other location in the genome. There are other concerns as well.
»A major limitation is that the off-target sites need to be previously identified for dRNAs to be designed against them. For novel gRNAs for which off-target sites have not yet been characterised, effective dRNAs cannot be easily designed,« explains Fowler.
An 'old' concept finds new uses
The term dead RNA or simply dRNA was first coined in 2015 where two research groups - Kiani et al. and Dahlman et al. - independently discovered that nuclease activity of Cas9 could be controlled by altering the length of the associated guide RNA. The two groups used the dRNAs for specific purposes that we will not elaborate on here, but Fowler recalls how their work led to the idea of dOTS:
»My colleague, John Rose, read the papers and wondered if Cas9 could be made to essentially compete with itself for target binding if the target sites of a dRNA and gRNA overlapped. He teamed up with Nicholas Popp to demonstrate that this Cas9 'self-competition' could be used to favourably bias editing outcomes.«
Rose and Popp are both first authors of the new paper, and they are not the only ones who have found creative uses of dRNAs to improve Cas9 editing efficiency. When Fowler and his team had sent their paper for review at Nature Communications and then uploaded the manuscript to BioRXiv, they were notified of a similar study carried out in Benjamin Taylor's group at AstraZeneca who published their results in August.
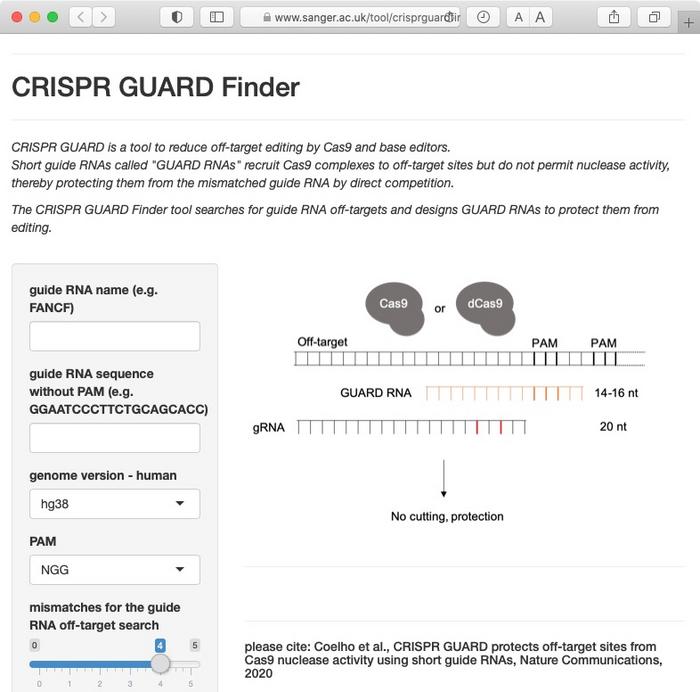
Taylor and his co-workers call their concept CRISPR GUARD (Guide RNA Assisted Reduction of Damage) and show that it can also support base editing. Moreover, they presented a free online tool to design GUARD RNAs.
Link to Fowler's original article in Nature Communications:
Suppression of unwanted CRISPR-Cas9 editing by co-administration of catalytically inactivating truncated guide RNAs
Tags
ArticleInterviewOff-targetQuality ControlBase editorsCRISPR-CasCas9dCas9
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







